Quickly Evaluate Captured Side Effects.
collateral 
R is great at automating a data analysis over many groups in a dataset, but errors, warnings and other side effects can stop it in its tracks. There’s nothing worse than returning after an hour (or a day!) and discovering that 90% of your data wasn’t analysed because one group threw an error.
With collateral, you can capture side effects like errors to prevent them from stopping execution. Once your analysis is done, you can see a tidy view of the groups that failed, the ones that returned a results, and the ones that threw warnings, messages or other output as they ran.
You can even filter or summarise a data frame based on these side effects to automatically continue analysis without failed groups (or perhaps to show a report of the failed ones).
For example, this screenshot shows a data frame that’s been nested using groups of the cyl column. The qlog column shows the results of an operation that’s returned results for all three groups but also printed a warning for one of them:
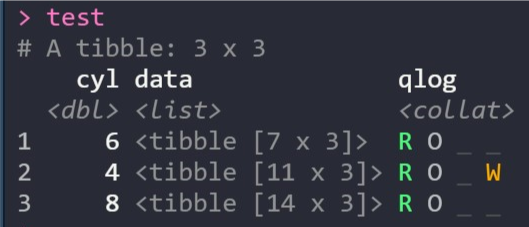
If you’re not familiar with purrr or haven’t used a list-column workflow in R before, the collateral vignette shows you how it works, the benefits for your analysis and how collateral simplifies the process of handling complex mapped operations.
If you’re already familiar with purrr, the tl;dr is that collateral::map_safely() and collateral::map_quietly() (and their map2 and pmap variants) will automatically wrap your supplied function in safely() or quietly() (or both: peacefully()) and will provide enhanced printed output and tibble displays. You can then use the has_*() and tally_*() functions to filter or summarise the returned tibbles or lists.
Installation
You can install the released version of collateral from CRAN with:
install.packages("collateral")
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("jimjam-slam/collateral")
Example
This example uses the famous mtcars dataset—but first, we’re going to sabotage a few of the rows by making them negative. The log function produces NaN with a warning when you give it a negative number.
It’d be easy to miss this in a non-interactive script if you didn’t explicitly test for the presence of NaN! Thankfully, with collateral, you can see which operations threw errors, which threw warnings, and which produced output:
library(tibble)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(tidyr)
library(collateral)
test <-
# tidy up and trim down for the example
mtcars %>%
rownames_to_column(var = "car") %>%
as_tibble() %>%
select(car, cyl, disp, wt) %>%
# spike some rows in cyl == 4 to make them fail
mutate(wt = dplyr::case_when(
wt < 2 ~ -wt,
TRUE ~ wt)) %>%
# nest and do some operations peacefully
nest(data = -cyl) %>%
mutate(qlog = map_peacefully(data, ~ log(.$wt)))
# look at the results
test
#> # A tibble: 3 × 3
#> cyl data qlog
#> <dbl> <list> <collat>
#> 1 6 <tibble [7 × 3]> R _ _ _ _
#> 2 4 <tibble [11 × 3]> R _ _ W _
#> 3 8 <tibble [14 × 3]> R _ _ _ _
Here, we can see that all operations produced output (because NaN is still output)—but a few of them also produced warnings! You can then see those warnings…
test %>% mutate(qlog_warning = map_chr(qlog, "warnings", .null = NA))
#> # A tibble: 3 × 4
#> cyl data qlog qlog_warning
#> <dbl> <list> <collat> <chr>
#> 1 6 <tibble [7 × 3]> R _ _ _ _ <NA>
#> 2 4 <tibble [11 × 3]> R _ _ W _ NaNs produced
#> 3 8 <tibble [14 × 3]> R _ _ _ _ <NA>
… filter on them…
test %>% filter(!has_warnings(qlog))
#> # A tibble: 2 × 3
#> cyl data qlog
#> <dbl> <list> <collat>
#> 1 6 <tibble [7 × 3]> R _ _ _ _
#> 2 8 <tibble [14 × 3]> R _ _ _ _
… or get a summary of them, for either interactive or non-interactive purposes:
summary(test$qlog)
#> 3 elements in total.
#> 3 elements returned results,
#> 3 elements delivered output,
#> 0 elements delivered messages,
#> 1 element delivered warnings, and
#> 0 elements threw an error.
Other features
The collateral package is now fully integrated with the furrr package, so you can safely and quietly iterate operations across CPUs cores or remote nodes. All collateral mappers have future_*-prefixed variants for this purpose.
Support
If you have a problem with collateral, please don’t hesitate to file an issue or contact me!
 ]
] 

