Distance Metrics for Image Color Similarity.
colordistance 1.1.1
Publication: https://peerj.com/articles/6398/
An R package with functions for quantifying the differences between colorful objects. Loads and displays images, selectively masks specified background colors, bins pixels by color using either data-dependent or automatically generated color bins, quantitatively measures color similarity among images using one of several distance metrics for comparing pixel color clusters, and clusters images by object color similarity. Originally written for use with organism coloration (reef fish color diversity, butterfly mimicry, etc), but easily applicable for any image set.
News
March 18, 2021: Minor updates to do with spatstat package (a sub-package, spatstat.geom, is now required instead). This shouldn't affect installation or usage.
November 11, 2020: Transparencies (alpha channel) can now be used to mask image backgrounds. By default, the presence of transparent pixels in a PNG overrides other background parameters, and the transparent pixels are ignored as background. This behavior can be disabled by setting alpha.channel = FALSE in any function that takes an image path as an argument. This allows users to specify the background in an image without having to decide on a background color that is sufficiently different from the object of interest. Backgrounds of uniform color can easily be rendered transparent using Photoshop, ImageJ, or GIMP.
February 19, 2020: Thanks to Evelyn Taylor-Cox for pointing out a bug with getLabHist when specifying a.bounds and b.bounds, resulting in bins that did not sum to 1. The bug should now be fixed in the development version!
February 6, 2019: Our methods paper for colordistance is out in PeerJ! Find it here: https://peerj.com/articles/6398/.
December 27, 2018: Fixed a bug when converting color clusters using convertColorSpace.
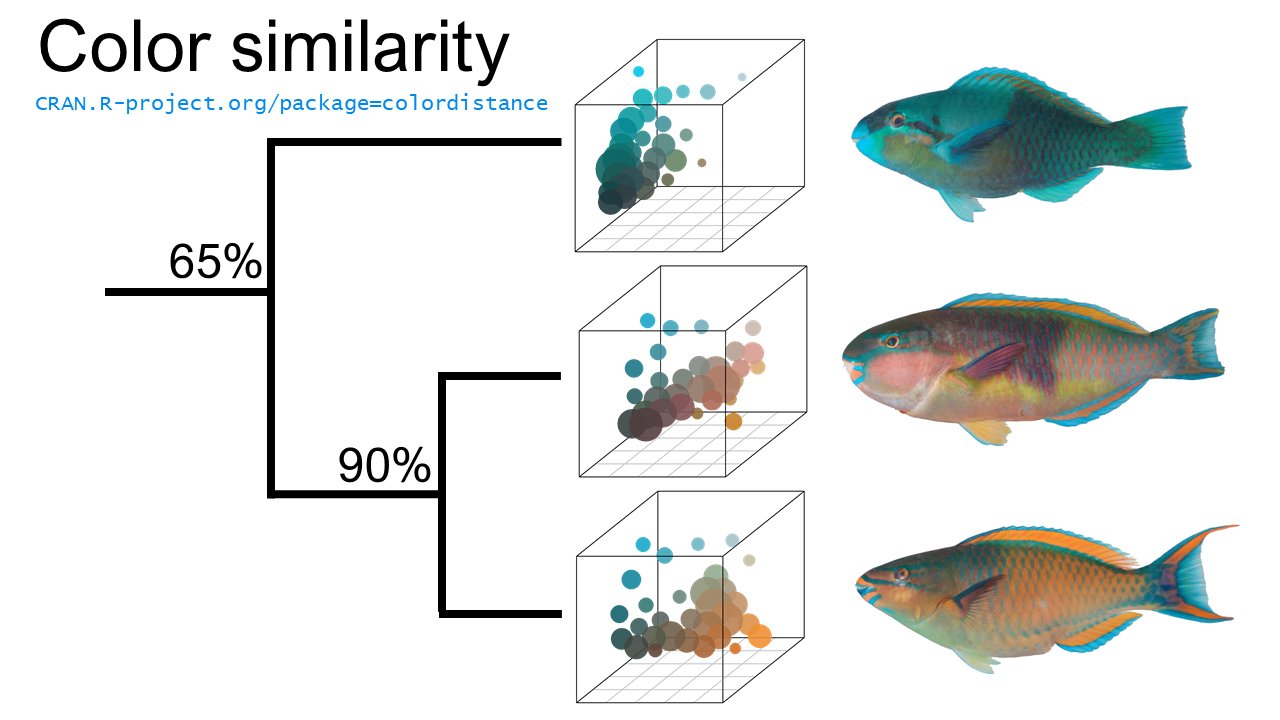
July 10, 2018: Added scatter3dclusters function to plot clusters in color space, scaled according to their size and colored according to their color. This is helpful for visualizing the distributions that colordistance actually compares to come up with a distance matrix, since the histograms can give the misleading impression that the clusters are treated as one-dimensional after binning. Also tweaked some compatibilities.
June 26, 2018: Added option to perform analyses CIELAB color space, as well as warnings about perceptual non-uniformity of RGB space. RGB (with warning) is still the default in order to prompt users to read up on CIELAB before using it. See "Color Spaces" and "CIELab Analyses" vignettes.
April 19, 2018: Functions for combining data across a set of images (combineClusters and combineList) added. Useful for pooling multiple images of the same individual, species, etc before analysis.
Input: Set(s) of JPEG or PNG images of colorful objects, optionally with backgrounds masked out.
Output: Color clusters, visualizations for color binning and image similarity, and distance matrices quantifying color similarity between images.
Requirements: R >= 3.3.2
Documentation: https://hiweller.github.io/colordistance/
Author: Hannah Weller
Contact: [email protected]
Installation
The development version of colordistance can be found at https://github.com/hiweller/colordistance.
To install the development version of colordistance in R:
Install the
devtoolspackage (install.packages("devtools")).Install
colordistancewithout vignettes (long-form documentation) to save time and space or with vignettes for offline access to help documents.# Without vignettes devtools::install_github("hiweller/colordistance") # With vignettes devtools::install_github("hiweller/colordistance", build_vignettes=TRUE)You can access help documents by running
help(package="colordistance")and clicking on the html files or, if you setbuild_vignettes=TRUEduring install, runvignette("colordistance-introduction").
To install the stable release version on CRAN (https://CRAN.R-project.org/package=colordistance), just run install.packages("colordistance").
Documentation
All of the colordistance vignettes that (optionally) come with the package are also available online at https://hiweller.github.io/colordistance/. I recommend reading at least the introduction before getting started.
Quickstart
To get started with colordistance, you'll need:
A set of images of objects you want to compare, ideally as consistent with each other as possible in terms of lighting and angle, and with anything you want to ignore masked out with a uniform background color. Need something to get started? Try these butterflyfish photos!
colordistancealso comes with an example set of Heliconius butterfly pictures from Meyer, 2006, which you can access viasystem.file("extdata", "Heliconius", package="colordistance")in R.R version 3.3.2 or later.
Estimates for the upper and lower RGB bounds for your background color. R reads in pixels channels with a 0-1 intensity range instead of the typical 0-255 (so pure red would be [1, 0, 0], green would be [0, 1, 0], blue would be [0, 0, 1], and so on). Background masking is rarely perfect, so you'll need to specify an upper and lower threshold for the background cutoff - around 0.2 usually does it. So if your background is white, your lower threshold would be [0.8, 0.8, 0.8] and your upper would be [1, 1, 1]. The default background color for
colordistanceis bright green, [0, 1, 0].
To run an analysis with all the default settings (bright green background masking, RGB color histograms with 3 bins per channel, and earth mover's distance for color distance metric -- see documentation), just run:
colordistance::imageClusterPipeline("path/to/images/folder")
You'll get a blue and yellow heatmap with a cluster dendrogram and labels taken from the image names. Yellow cells correspond to dissimilar images; blue cells correspond to more similar images. If those scores don't look right, try changing the number of bins (bins argument), the distance metric (distanceMethod argument), and making sure you're masking out the right background color.
Citing colordistance
Our methods paper describing colordistance is now out in PeerJ! Please cite the package as: Weller HI, Westneat MW. 2019. Quantitative color profiling of digital images with earth mover’s distance using the R package colordistance. PeerJ 7:e6398 https://doi.org/10.7717/peerj.6398
Contributions and bug reports
If something is wrong or confusing, or if you'd like to see a change, please create an issue on the issues page of the GitHub repository, as it allows other people to see it. You can also email me at [email protected].
If you would like to contribute, feel free to make a pull request or email me with your thoughts.
Questions?
Email me at [email protected]. I generally respond within 48 hours.