Description
Conditional Formatting in Data Frames.
Description
Apply and visualize conditional formatting to data frames in R. It renders a data frame with cells formatted according to criteria defined by rules, using a tidy evaluation syntax. The table is printed either opening a web browser or within the 'RStudio' viewer if available. The conditional formatting rules allow to highlight cells matching a condition or add a gradient background to a given column. This package supports both 'HTML' and 'LaTeX' outputs in 'knitr' reports, and exporting to an 'xlsx' file.
README.md
Introduction to condformat
Sergio Oller 2023-10-08
condformat renders a data frame in which cells in columns are formatted according to several rules or criteria.
Browse source code
Checkout the code and browse it at https://github.com/zeehio/condformat.
How to install condformat:
Dependencies
If you want to use the PDF output you will need the xcolor LaTeX package. Either use the full texlive distribution, or install latex-xcolor on Debian and derivatives.
Package installation
From CRAN:
install.packages("condformat")To install the latest development version:
remotes::install_github("zeehio/condformat")
Example
data(iris)
library(condformat)
condformat(iris[c(1:5,70:75, 120:125),]) %>%
rule_fill_discrete(Species) %>%
rule_fill_discrete(c(Sepal.Width, Sepal.Length),
expression = Sepal.Width > Sepal.Length - 2.25,
colours = c("TRUE" = "#7D00FF")) %>%
rule_fill_gradient2(Petal.Length) %>%
rule_text_bold(c(Sepal.Length, Species), Species == "versicolor") %>%
rule_text_color(Sepal.Length,
expression = ifelse(Species == "setosa", "yellow", "")) %>%
rule_fill_bar(Petal.Width, limits = c(0, NA)) %>%
theme_grob(rows = NULL) %>%
condformat2grob()
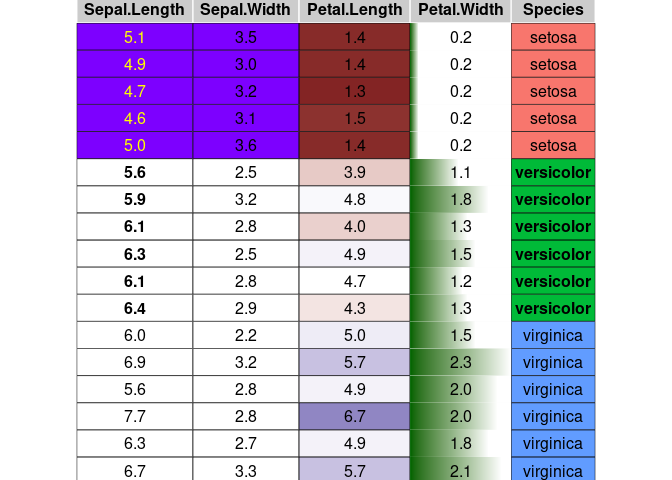
This example covers most of the condformat rules.
Rules and output engine support
| Rule | HTML | LaTeX | gtable | Excel |
|---|---|---|---|---|
rule_fill_discrete | X | X | X | X |
rule_fill_gradient | X | X | X | X |
rule_fill_gradient2 | X | X | X | X |
rule_text_color | X | X | X | X |
rule_text_bold | X | X | X | X |
rule_fill_bar | X | X | ||
rule_css | X. |
