R Binding for the 'Certifiably Optimal RulE ListS (Corels)'.
corels: R interface to 'Certifiably Optimal RulE ListS' (Corels)
What is it?
CORELS is a custom discrete optimization technique for building rule lists over a categorical feature space. The algorithm provides the optimal solution with a certificate of optimality. By leveraging algorithmic bounds, efficient data structures, and computational reuse, it achieves several orders of magnitude speedup in time and a massive reduction of memory consumption. This approach produces optimal rule lists on practical problems in seconds, and offers a novel alternative to CART and other decision tree methods.
See the C++ implementation, the live website, the Python implementation, the arXiv paper, the JMLR paper, the senior thesis or the KDD 2017 paper for more.
More about Corels can also be read in this recent post at The Morning Paper.
Illustration
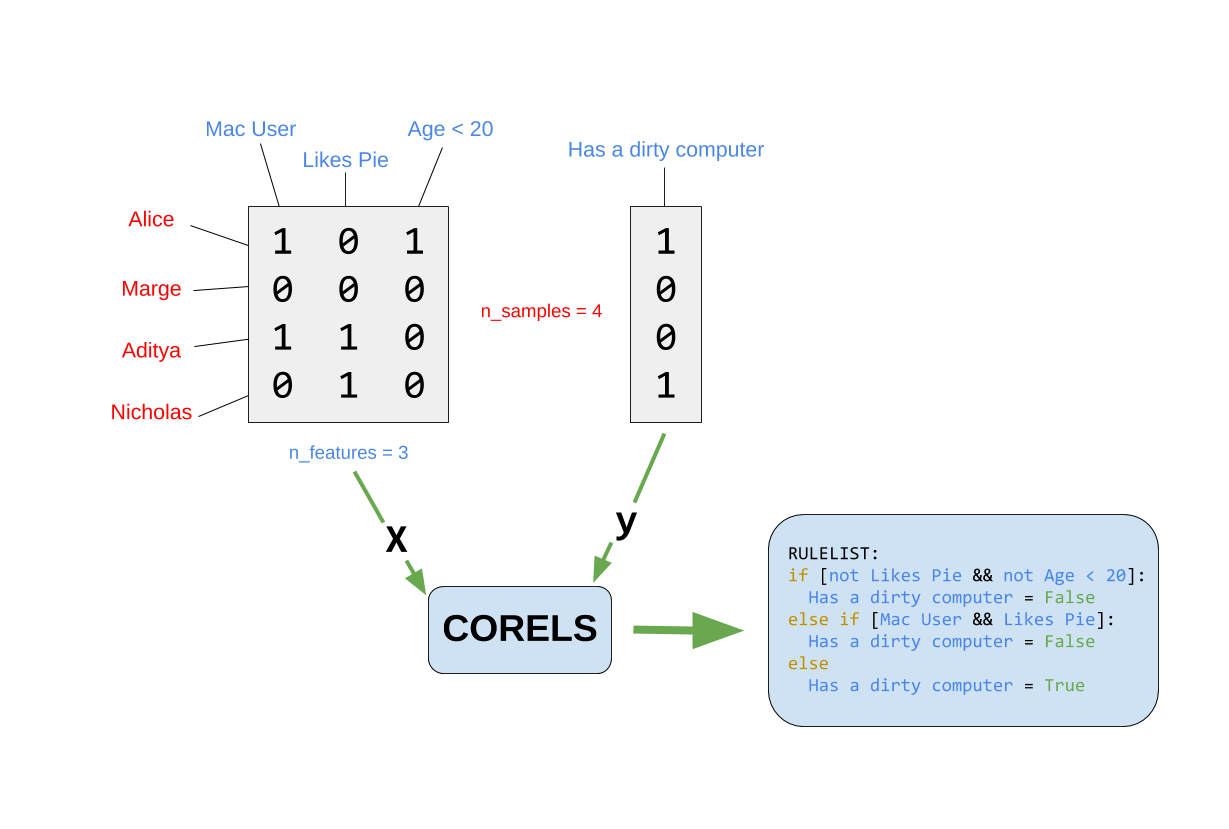
With thanks to the Python implementation for the image.
What is this package?
We use Rcpp to connect the Corels C++ implementation to R.
Status
Installs and works fine, and passed R CMD check. Several extensions possible, see below.
Installation
As the package is not (yet?) on CRAN, do
remotes::install_github("eddelbuettel/rcppcorels")
Note of the GNU GMP library is now optional; configure will enable (via a -DGMP define and link instructions) if found. GMP will improve performance, so you may want to do sudo apt-get install libgmp-dev, or whatever equivalent command you need to install it on your system.
TODOs
Plenty such as adding Travis CI support, adding configure code to detect GNU GMP presence, adding examples, factoring out (input) data reader code, possibly visualizing decision trees, and more.
Author
Dirk Eddelbuettel wrote the R package and integration.
Nicholas Larus-Stone and Elaine Angelino wrote the C++ implementation of Corels.
Elaine Angelino, Nicholas Larus-Stone, Daniel Alabi, Margo Seltzer, and Cynthia Rudin wrote the paper.
Corels uses the rulelib library by Yang et al described in the 2016 arXiv paper by Hongyu Yang, Cynthia Rudin, and Margo Seltzer with this code repo and in the 2015 arXiv paper by Benjamin Letham, Cynthia Rudin, Tyler H. McCormick and David Madigan now published in Annals of Statistics.
License
This package is released under the GPL-3, as is Corels.
The rulelib library is released under the MIT license.