COVID-19 Cases in Switzerland and Principality of Liechtenstein.
covid19swiss
The covid19swiss R package provides a tidy format dataset of the 2019 Novel Coronavirus COVID-19 (2019-nCoV) pandemic outbreak in Switzerland cantons and the Principality of Liechtenstein (FL). The package is following the data structure of the Covid19R project.
The covid19swiss dataset includes the following fields:
date- the timestamp of the case, aDateobjectlocation- the Cantons of Switzerland and the Principality of Liechtenstein (FL) abbreviation codelocation_type- description of the location, either Canton of Switzerland or the Principality of echtensteinlocation_code- a canton index code for merging geometry data from the rnaturalearth package, ailable only for Switzerland cantonslocation_code_type- the name of code in the rnaturalearth package for Switzerland mapdata_type- the type of casevalue- the number of cases corresponding to thedateanddata_typefields
Where the available data_type field includes the following cases:
tested_total- cumulative number of tests performed as of the datecases_total- cumulative confirmed Covid-19 cases as of the current datehosp_new- new hospitalizations on the current datehosp_current- current number of hospitalized patients as of the current dateicu_current- number of hospitalized patients in ICUs as of the current datevent_current- number of hospitalized patients requiring ventilation as of the current daterecovered_total- cumulative number of patients recovered as of the current datedeaths_total- cumulative deaths due to Covid-19 as of the current date
More information can be found the following vignettes:
- Introduction to the covid19swiss Dataset
- Updating the covid19swiss Dataset
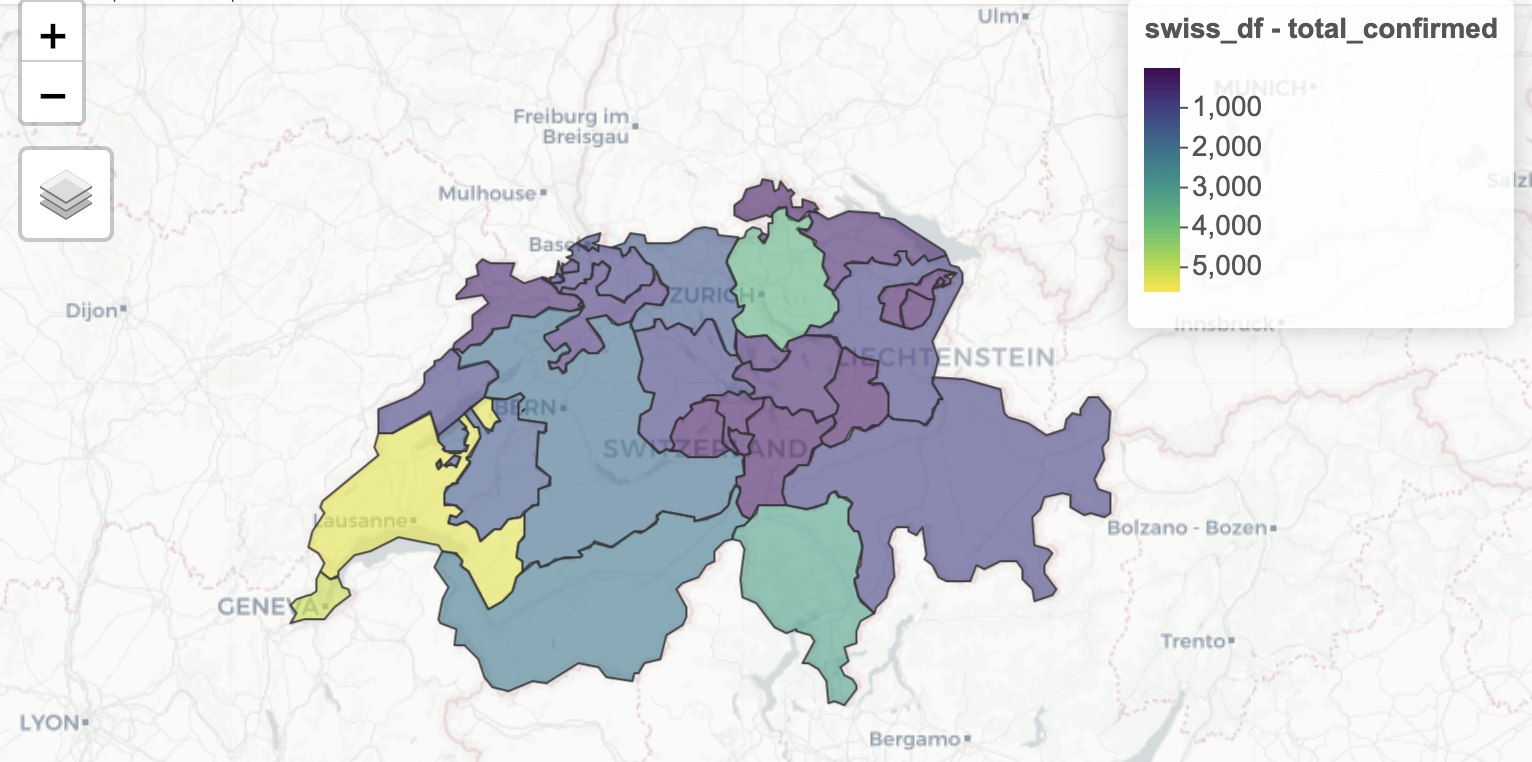
- Geospatial Visualization of Switzerland Cantons (non CRAN vignette)
Data source: Specialist Unit for Open Government Data Canton of Zurich, raw data is available on the following repository. Special thanks for all the people that collaborate and contribute to pull the data from multiple sources and make this data available!
Installation
You can install the released version of covid19swiss from CRAN with:
install.packages("covid19swiss")
Or, install the most recent version from GitHub with:
# install.packages("devtools")
devtools::install_github("Covid19R/covid19swiss")
Data refresh
The covid19swiss package dev version is been updated on a daily bases. The update_swiss_data function enables a simple refresh of the installed package datasets with the most updated version on Github:
library(covid19swiss)
update_swiss_data()
More information about updating the data is available on this vignette
Note: must restart the R session after using the update_swiss_data function in order to have the updates available
Usage
data(covid19swiss)
head(covid19swiss)
#> date location location_type location_code location_code_type data_type value
#> 1 2020-01-24 GE Canton of Switzerland CH.GE gn_a1_code tested_total 4
#> 2 2020-01-24 GE Canton of Switzerland CH.GE gn_a1_code cases_total NA
#> 3 2020-01-24 GE Canton of Switzerland CH.GE gn_a1_code hosp_new NA
#> 4 2020-01-24 GE Canton of Switzerland CH.GE gn_a1_code hosp_current NA
#> 5 2020-01-24 GE Canton of Switzerland CH.GE gn_a1_code icu_current NA
#> 6 2020-01-24 GE Canton of Switzerland CH.GE gn_a1_code vent_current NA
Wide format
library(tidyr)
covid19swiss_wide <- covid19swiss %>%
pivot_wider(names_from = data_type, values_from = value)
head(covid19swiss_wide)
#> # A tibble: 6 x 13
#> date location location_type location_code location_code_type tested_total cases_total hosp_new hosp_current icu_current vent_current recovered_total deaths_total
#> <date> <chr> <chr> <chr> <chr> <int> <int> <int> <int> <int> <int> <int> <int>
#> 1 2020-01-24 GE Canton of Switzerland CH.GE gn_a1_code 4 NA NA NA NA NA NA NA
#> 2 2020-01-25 GE Canton of Switzerland CH.GE gn_a1_code 8 NA NA NA NA NA NA NA
#> 3 2020-01-26 GE Canton of Switzerland CH.GE gn_a1_code 11 NA NA NA NA NA NA NA
#> 4 2020-01-27 GE Canton of Switzerland CH.GE gn_a1_code 18 NA NA NA NA NA NA NA
#> 5 2020-01-28 GE Canton of Switzerland CH.GE gn_a1_code 27 NA NA NA NA NA NA NA
#> 6 2020-01-29 GE Canton of Switzerland CH.GE gn_a1_code 54 NA NA NA NA NA NA NA
Missing values
The data is collected from multiple resources on the canton level, and not necessarily each data resource provides the same field. Therefore, some fields, such as total recovered or total tested may not be available at this point for some cantons and are marked as missing values (i.e., NA).
Contributing
Please submit issues and pull requests with any package improvements!
Please note that the ‘covid19swiss’ project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.


