Description
Cases of COVID-19 in Tunisia.
Description
Data personally collected about the spread of COVID-19 (SARS-COV-2) in Tunisia <https://github.com/MounaBelaid/covid19datatunisia>.
README.md
This package provides per-day data on COVID-19 cases in Tunisia.
The data is an average over all of the sources provided by Mouna Belaid. Please check out this link.
Installation
Using the programming language R, you can install the released version of covid19tunisia with :
remotes::install_github("MounaBelaid/covid19tunisia")
Usage
data <- refresh_covid19tunisia()
head(data)
#> # A tibble: 6 x 7
date location location_type location_code location_code_type data_type value
<date> <chr> <chr> <chr> <chr> <chr> <dbl>
1 2020-03-02 Gafsa state TN-71 iso_3166_2 cases_new 1
2 2020-03-08 Mahdia state TN-53 iso_3166_2 cases_new 1
3 2020-03-09 Bizerte state TN-23 iso_3166_2 cases_new 1
4 2020-03-09 Mahdia state TN-53 iso_3166_2 cases_new 1
5 2020-03-09 Tunis state TN-11 iso_3166_2 cases_new 1
6 2020-03-10 Mahdia state TN-53 iso_3166_2 cases_new 1
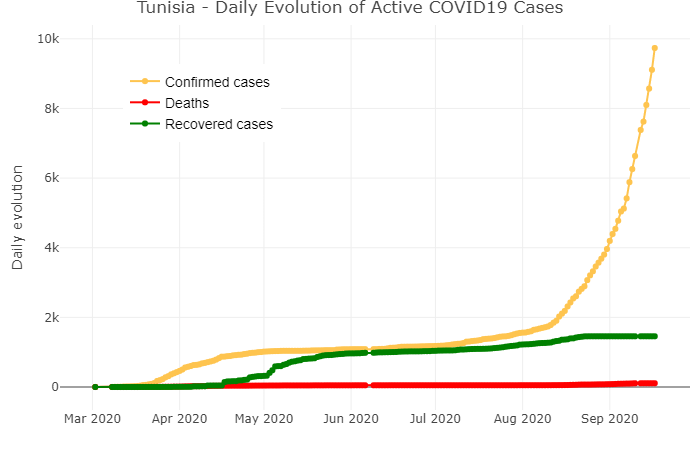
Plotting the daily evolution of active cases
# Transform the data
library(dplyr)
library(tidyr)
library(plotly)
data_transformed <- data %>% group_by(date, data_type) %>% summarise(value = sum(value)) %>%
spread(data_type, value)
head(data_transformed)
# A tibble: 6 x 4
# Groups: date [6]
date cases_new deaths_new recovered_new
<date> <dbl> <dbl> <dbl>
1 2020-03-02 1 0 0
2 2020-03-08 1 0 0
3 2020-03-09 3 0 0
4 2020-03-10 1 0 0
5 2020-03-11 1 0 0
6 2020-03-12 6 0 0
data_transformed %>%
ungroup() %>% plot_ly(type = 'scatter',
mode = 'lines + markers')%>%
add_trace(x = ~date, y = ~cumsum(cases_new),
name = 'Confirmed cases',
marker = list(color = '#fec44f'),
line = list(color = '#fec44f'),
hoverinfo = "text",
text = ~paste(cases_new, "New confirmed cases\n", cumsum(cases_new), 'Total number of infected cases on', date)) %>%
add_trace(x = ~date, y = ~cumsum(deaths_new),
name = 'Deaths',
marker = list(color = 'red'),
line = list(color = 'red'),
hoverinfo = "text",
text = ~paste(deaths_new, "New deaths\n", cumsum(deaths_new), 'Total number of deaths on', date)) %>%
add_trace(x = ~date, y = ~cumsum(recovered_new),
name = 'Recovered cases',
marker = list(color = 'green'),
line = list(color = 'green'),
hoverinfo = "text",
text = ~paste(recovered_new, "New recovered cases\n", cumsum(recovered_new), 'Total number of recovered cases on', date)) %>%
layout(title = 'Tunisia - Daily Evolution of Active COVID19 Cases',
legend = list(x = 0.1, y = 0.9,
font = list(family = "sans-serif", size = 14, color = "#000"), bgcolor = "",
bordercolor = "#FFFFFF", borderwidth = 2),
xaxis = list(title = ""),
yaxis = list(side = 'left', title = 'Daily evolution', showgrid = TRUE, zeroline = TRUE))
Supporting Shiny Application
A supporting Shiny Application for the covid19tunisia dataset available here.