Cases of COVID-19 in the United States.
covid19us
This is an R wrapper around the COVID Tracking Project API. It provides updates on the spread of the virus in the US with a few simple functions.
Installation
install.packages("covid19us")
Or the dev version:
devtools::install_github("aedobbyn/covid19us")
Examples
library(covid19us)
Get the most recent COVID-19 top-line data for the country:
get_us_current()
#> # A tibble: 1 x 18
#> positive negative pending hospitalized_cu… hospitalized_cu… in_icu_currently
#> <int> <int> <int> <int> <int> <int>
#> 1 1520778 10713209 2944 41174 159634 9829
#> # … with 12 more variables: in_icu_cumulative <int>,
#> # on_ventilator_currently <int>, on_ventilator_cumulative <int>,
#> # recovered <int>, hash <chr>, last_modified <chr>, death <int>,
#> # hospitalized <int>, total <int>, total_test_results <int>, notes <chr>,
#> # request_datetime <dttm>
Or the same by state:
get_states_current()
#> # A tibble: 56 x 30
#> state positive positive_score negative_score negative_regula…
#> <chr> <int> <int> <int> <int>
#> 1 AK 399 1 1 1
#> 2 AL 12376 1 1 0
#> 3 AR 4923 1 1 1
#> 4 AZ 14566 1 1 0
#> 5 CA 81795 1 1 0
#> 6 CO 22202 1 1 1
#> 7 CT 38430 1 1 1
#> 8 DC 7434 1 1 1
#> 9 DE 8037 1 1 1
#> 10 FL 46944 1 1 1
#> # … with 46 more rows, and 25 more variables: commercial_score <int>,
#> # grade <chr>, score <int>, notes <chr>, data_quality_grade <chr>,
#> # negative <int>, pending <int>, hospitalized_currently <int>,
#> # hospitalized_cumulative <int>, in_icu_currently <int>,
#> # in_icu_cumulative <int>, on_ventilator_currently <int>,
#> # on_ventilator_cumulative <int>, recovered <int>, last_update <dttm>,
#> # check_time <dttm>, death <int>, hospitalized <int>, total <int>,
#> # total_test_results <int>, fips <chr>, date_modified <dttm>,
#> # date_checked <dttm>, hash <chr>, request_datetime <dttm>
Daily state counts can be filtered by state and/or date:
get_states_daily(
state = "NY",
date = "2020-03-17"
)
#> # A tibble: 1 x 27
#> date state positive negative pending hospitalized_cu… hospitalized_cu…
#> <date> <chr> <int> <int> <int> <int> <int>
#> 1 2020-03-17 NY 1700 5506 NA 325 NA
#> # … with 20 more variables: in_icu_currently <int>, in_icu_cumulative <int>,
#> # on_ventilator_currently <int>, on_ventilator_cumulative <int>,
#> # recovered <int>, data_quality_grade <chr>, last_update <dttm>, hash <chr>,
#> # date_checked <dttm>, death <int>, hospitalized <int>, total <int>,
#> # total_test_results <int>, fips <chr>, death_increase <int>,
#> # hospitalized_increase <int>, negative_increase <int>,
#> # positive_increase <int>, total_test_results_increase <int>,
#> # request_datetime <dttm>
For data in long format:
(dat <- refresh_covid19us())
#> # A tibble: 80,123 x 7
#> date location location_type location_code location_code_t… data_type
#> <date> <chr> <chr> <chr> <chr> <chr>
#> 1 2020-05-19 AK state 02 fips_code positive
#> 2 2020-05-19 AK state 02 fips_code negative
#> 3 2020-05-19 AK state 02 fips_code pending
#> 4 2020-05-19 AK state 02 fips_code hospital…
#> 5 2020-05-19 AK state 02 fips_code hospital…
#> 6 2020-05-19 AK state 02 fips_code in_icu_c…
#> 7 2020-05-19 AK state 02 fips_code in_icu_c…
#> 8 2020-05-19 AK state 02 fips_code on_venti…
#> 9 2020-05-19 AK state 02 fips_code on_venti…
#> 10 2020-05-19 AK state 02 fips_code recovered
#> # … with 80,113 more rows, and 1 more variable: value <int>
Which can be easier to plot
library(dplyr)
library(ggplot2)
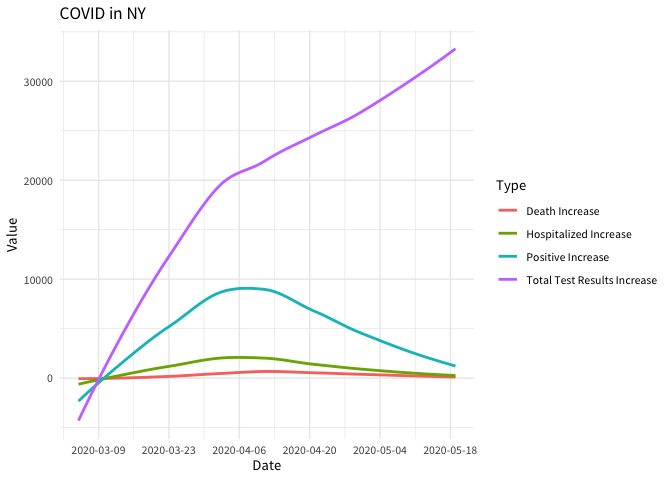
dat %>%
filter(
location == "NY" &
data_type %in%
c(
"positive_increase",
"total_test_results_increase",
"death_increase",
"hospitalized_increase"
)
) %>%
mutate(
Type = data_type %>%
stringr::str_replace_all("_", " ") %>%
stringr::str_to_title()
) %>%
arrange(date) %>%
ggplot(aes(date, value, color = Type)) +
geom_smooth(se = FALSE) +
scale_x_date(date_breaks = "2 weeks") +
labs(title = "COVID in NY") +
xlab("Date") +
ylab("Value") +
theme_minimal(base_family = "Source Sans Pro")
To get information about the data:
get_info_covid19us()
#> # A tibble: 1 x 10
#> data_set_name package_name function_to_get… data_details data_url license_url
#> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 covid19us covid19us refresh_covid19… Open Source… https:/… https://gi…
#> # … with 4 more variables: data_types <chr>, location_types <chr>,
#> # spatial_extent <chr>, has_geospatial_info <lgl>
All Functions
get_counties_info
get_info_covid19us
get_states_current
get_states_daily
get_states_info
get_tracker_urls
get_us_current
get_us_daily
refresh_covid19us
Other Details
All of the data sources can be found with
get_tracker_urls()- The
filtercolumn gives information about how the COVID Tracking Project’s scraper currently scrapes data from the page (xpaths, CSS selectors, functions used, etc.)
- The
State breakdowns include DC as well as some US territories including American Samoa (AS), Guam (GU), Northern Mariana Islands (MP), Puerto Rico (PR), and the Virgin Islands (VI)
Acronyms
- PUI: persons under investigation
- PUM: persons under monitoring (one step before PUI)
Time zone used is Eastern Standard Time
PRs and bug reports / feature requests welcome. Stay safe!
