Discriminant Adaptive Nearest Neighbor Classification.
DANN
An implementation of Hastie and Tibshirani’s Discriminant Adaptive Nearest Neighbor Classification in R.
Package Introduction
In k nearest neighbors, the shape of the neighborhood is usually circular. Discriminant Adaptive Nearest Neighbor (dann) is a variation of k nearest neighbors where the shape of the neighborhood is data driven. The neighborhood is elongated along class boundaries and shrunk in the orthogonal direction. See Discriminate Adaptive Nearest Neighbor Classification by Hastie and Tibshirani.
Models:
- dann
- sub-dann
This package implements DANN and sub-DANN in section 4.1 of the publication and is based on Christopher Jenness’s python implementation.
Example 1: dann
Arguments:
- k - The number of points in the neighborhood. Identical to k in standard k nearest neighbors.
- neighborhood_size - The number of data points used to estimate a good shape for the neighborhood.
- epsilon - Softening parameter. Usually has the least affect on performance.
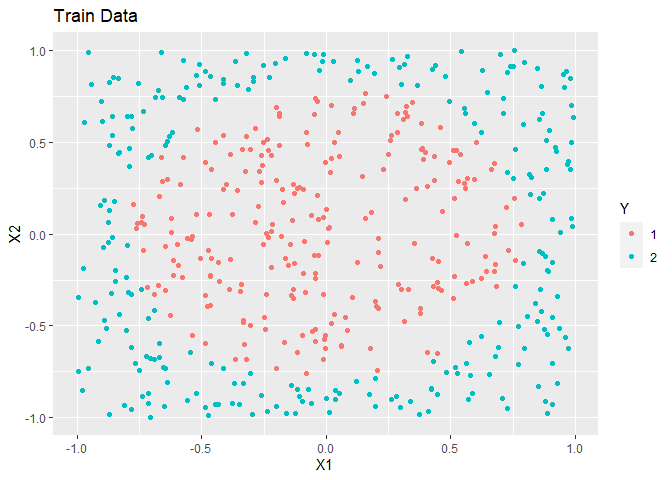
In this example, simulated data is made. The overall trend is a circle inside a square.
library(dann)
library(dplyr, warn.conflicts = FALSE)
library(ggplot2)
library(mlbench)
set.seed(1)
# Create training data
train <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(train) <- c("X1", "X2", "Y")
train <- train %>%
mutate(Y = as.numeric(Y))
ggplot(train, aes(x = X1, y = X2, colour = as.factor(Y))) +
geom_point() +
labs(title = "Train Data", colour = "Y")
To train a model, call dann.
model <- dann(formula = Y ~ X1 + X2, data = train, k = 5, neighborhood_size = 50, epsilon = 1)
To get class predictions, call predict with type equal to “class”
# Create test data
test <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(test) <- c("X1", "X2", "Y")
test <- test %>%
mutate(Y = as.numeric(Y))
yhat <- predict(object = model, new_data = test, type = "class")
yhat
#> # A tibble: 500 × 1
#> .pred_class
#> <fct>
#> 1 2
#> 2 1
#> 3 1
#> 4 2
#> 5 2
#> 6 2
#> 7 1
#> 8 1
#> 9 2
#> 10 1
#> # ℹ 490 more rows
To get probabilities, call predict with type equal to “prob”
yhat <- predict(object = model, new_data = test, type = "prob")
yhat
#> # A tibble: 500 × 2
#> .pred_1 .pred_2
#> <dbl> <dbl>
#> 1 0 1
#> 2 0.6 0.4
#> 3 1 0
#> 4 0 1
#> 5 0 1
#> 6 0 1
#> 7 1 0
#> 8 1 0
#> 9 0 1
#> 10 1 0
#> # ℹ 490 more rows
Example 2: sub_dann
In general, dann will struggle as unrelated variables are intermingled with informative variables. To deal with this, sub_dann projects the data onto a unique subspace and then calls dann on the subspace.
Arguments
- k - The number of points in the neighborhood. Identical to k in standard k nearest neighbors.
- neighborhood_size - The number of data points used to estimate a good shape for the neighborhood.
- epsilon - Softening parameter. Usually has the least affect on performance.
- weighted - Should the individual between class covariance matrices be weighted? FALSE corresponds to original publication.
- sphere - Type of covariance matrix to calculate.
- numDim - The number of dimensions to project the predictor variables on to.
In the below example there are 2 related variables and 5 that are unrelated. Will sub_dann to better than dann?
######################
# Circle data with unrelated variables
######################
set.seed(1)
train <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(train)[1:3] <- c("X1", "X2", "Y")
# Add 5 unrelated variables
train <- train %>%
mutate(
U1 = runif(500, -1, 1),
U2 = runif(500, -1, 1),
U3 = runif(500, -1, 1),
U4 = runif(500, -1, 1),
U5 = runif(500, -1, 1)
)
test <- mlbench.circle(500, 2) %>%
tibble::as_tibble()
colnames(test)[1:3] <- c("X1", "X2", "Y")
# Add 5 unrelated variables
test <- test %>%
mutate(
U1 = runif(500, -1, 1),
U2 = runif(500, -1, 1),
U3 = runif(500, -1, 1),
U4 = runif(500, -1, 1),
U5 = runif(500, -1, 1)
)
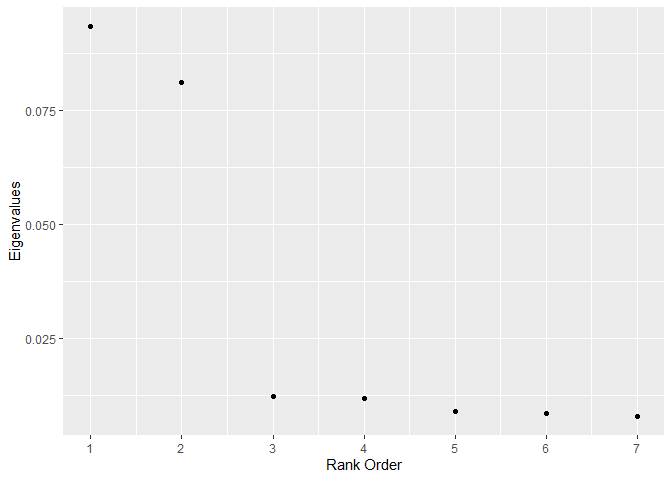
For sub_dann, the dimension of the subspace should be chosen based on the number of large eigenvalues. The graph suggests 2 (the correct answer).
graph_eigenvalues(
formula = Y ~ X1 + X2 + U1 + U2 + U3 + U4 + U5,
data = train,
neighborhood_size = 50,
weighted = FALSE,
sphere = "mcd"
)
dann_model <- dann(
formula = Y ~ X1 + X2 + U1 + U2 + U3 + U4 + U5,
data = train,
k = 3,
neighborhood_size = 50,
epsilon = 1
)
# numDim based on large eigenvalues
# weighted, sphere, and neighborhood_size kept consistent between sub_dann and graph_eigenvalues
sub_dann_model <- sub_dann(
formula = Y ~ X1 + X2 + U1 + U2 + U3 + U4 + U5,
data = train,
k = 3,
neighborhood_size = 50,
epsilon = 1,
weighted = FALSE,
sphere = "mcd",
numDim = 2
)
As expected, the dann model using all features does not fit the data well.
library(yardstick)
dann_yhat <- predict(object = dann_model, new_data = test, type = "prob")
dann_yhat <- test %>%
select(Y) %>%
bind_cols(dann_yhat)
roc_auc(data = dann_yhat, truth = Y, event_level = "first", .pred_1)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.725
sub_dann provides a major improvement.
sub_dann_yhat <- predict(object = sub_dann_model, new_data = test, type = "prob")
sub_dann_yhat <- test %>%
select(Y) %>%
bind_cols(sub_dann_yhat)
roc_auc(data = sub_dann_yhat, truth = Y, event_level = "first", .pred_1)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.935