Interactive and Reproducible Data Cleaning.
datacleanr 
datacleanr is a flexible and efficient tool for interactive data cleaning, and is inherently interoperable, as it seamlessly integrates into reproducible data analyses pipelines in R.
It can deal with nested tabular, as well as spatial and time series data.
Installation
The latest release on CRAN can be installed using:
install.packages("datacleanr")
You can install the development version of datacleanr with:
remotes::install_github("the-hull/datacleanr")
If you are using macOS, please make sure you have XQuartz installed, especially if you’ve recently updated your system.See these instructions here: https://CRAN.R-project.org/bin/macosx/
**In case the package installation fails due to a compilation error from magick, please try installing the dev headers for magick with sudo apt-get install libmagick++-dev. Also see this gh issue.
Design
datacleanr is developed using the shiny package, and relies on informative summaries, visual cues and interactive data selection and annotation. All data-altering operations are documented, and converted to valid R code (reproducible recipe), that can be copied, sent to an active RStudio script, or saved to disk.
There are four tabs in the app for these tasks:
- Set-up & Overview: define nesting structure based on (multiple) groups.
- Filtering: use
Rexpression to filter/subset data. - Visual Cleaning and Annotating: generate bivarirate (time series) plots and maps, as well as highlight and annotate individual observations. Cycle through nested groups to expedite exploration and cleaning. Histograms of original vs. ‘cleaned’ data can be generated.
- Extract: generate reproducible recipe and define outputs.
dcr_appalso returns all intermediate and final outputs invisibly to the activeRsession for later use (e.g. when batch processing)
Note, maps require columns lon and lat (X and Y) in decimal degrees in the data set to render.
Additional features
- Grouping: the grouping defined in the “Set-up and Overview” tab is carried forward through the app. These groups can be used to cycle through nested/granular data, and considerably speed up exploration and cleaning. These groups are also available for filtering (Filtering tab), where filter expressions can be scoped to group level (i.e. no groups, individual, all groups).
- Interoperability: when a logical (
TRUE\`FALSE) column named.dcrflag` is present, corresponding observations are rendered with different symbols in plots and maps. Use this feature to validate or cross-check external quality control or outlier flagging methods. - Batching: If data sets are too large, or too deeply nested (e.g. individual, plot, site, region, etc.), we recommend a split-combine approach to expedite the processing.
# prepare data into species sub-sets
iris_split <- split(x = iris,
f = iris$Species)
# run for each species
dcr_iris <- lapply(iris_split,
function(split){
datacleanr::dcr_app(split)
})
Getting started
The documentation for (?dcr_app()) explains the basic use and all features. Throughout the app, there are conveniently-placed help links that provide details on features.
Demonstration
Launch datacleanr’s interactive app with dcr_app(). The following examples demonstrate basic use and highlight features across the four app tabs.
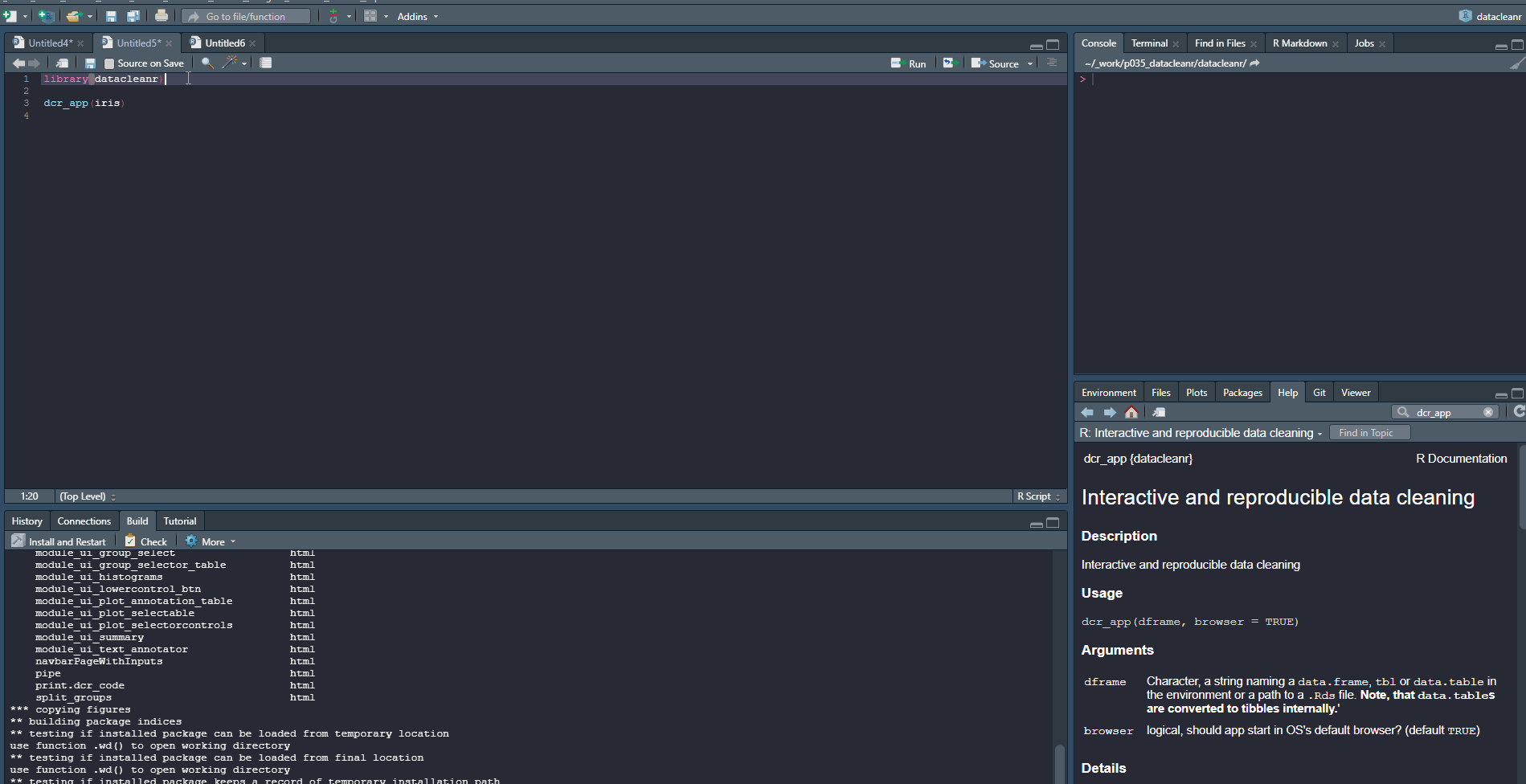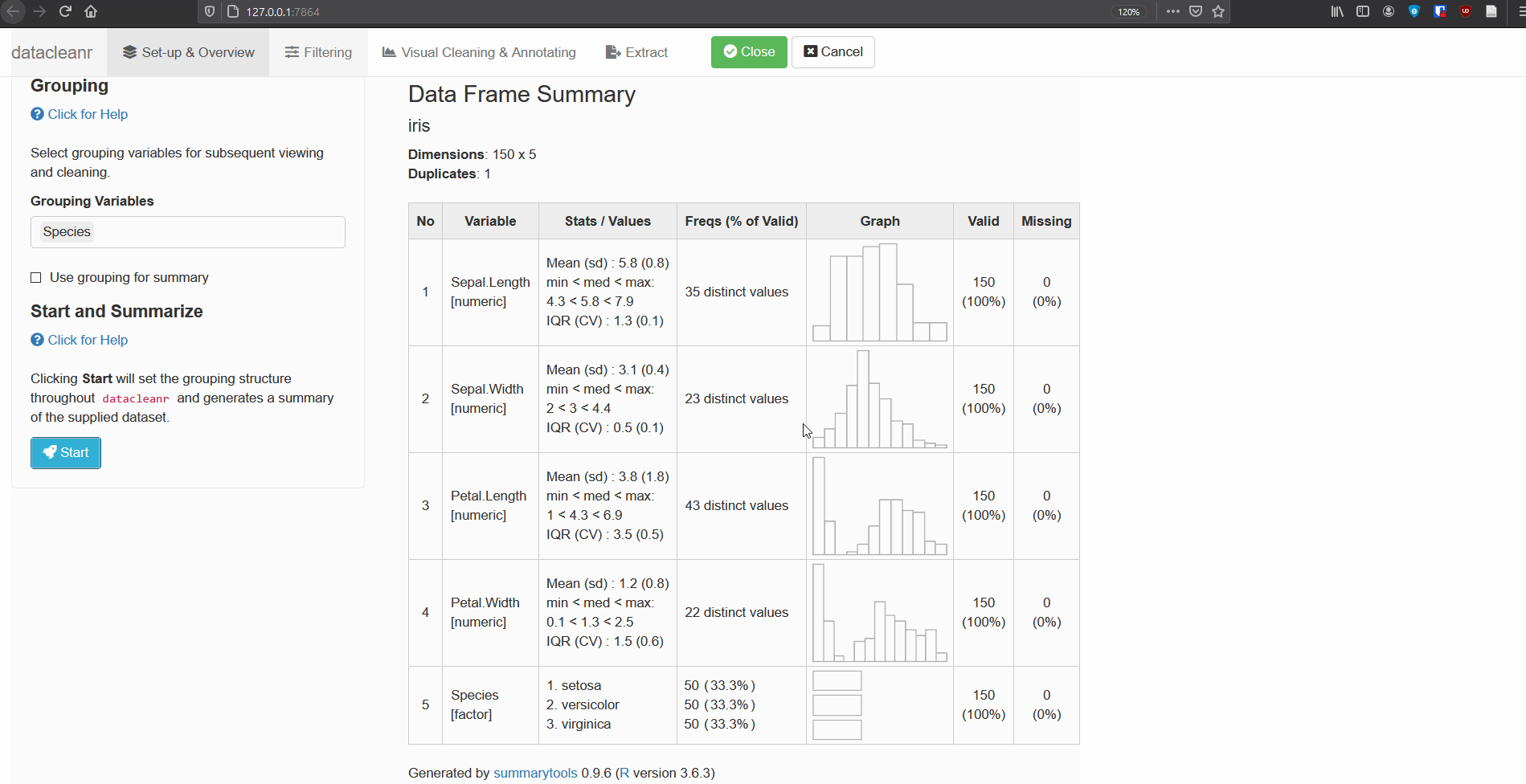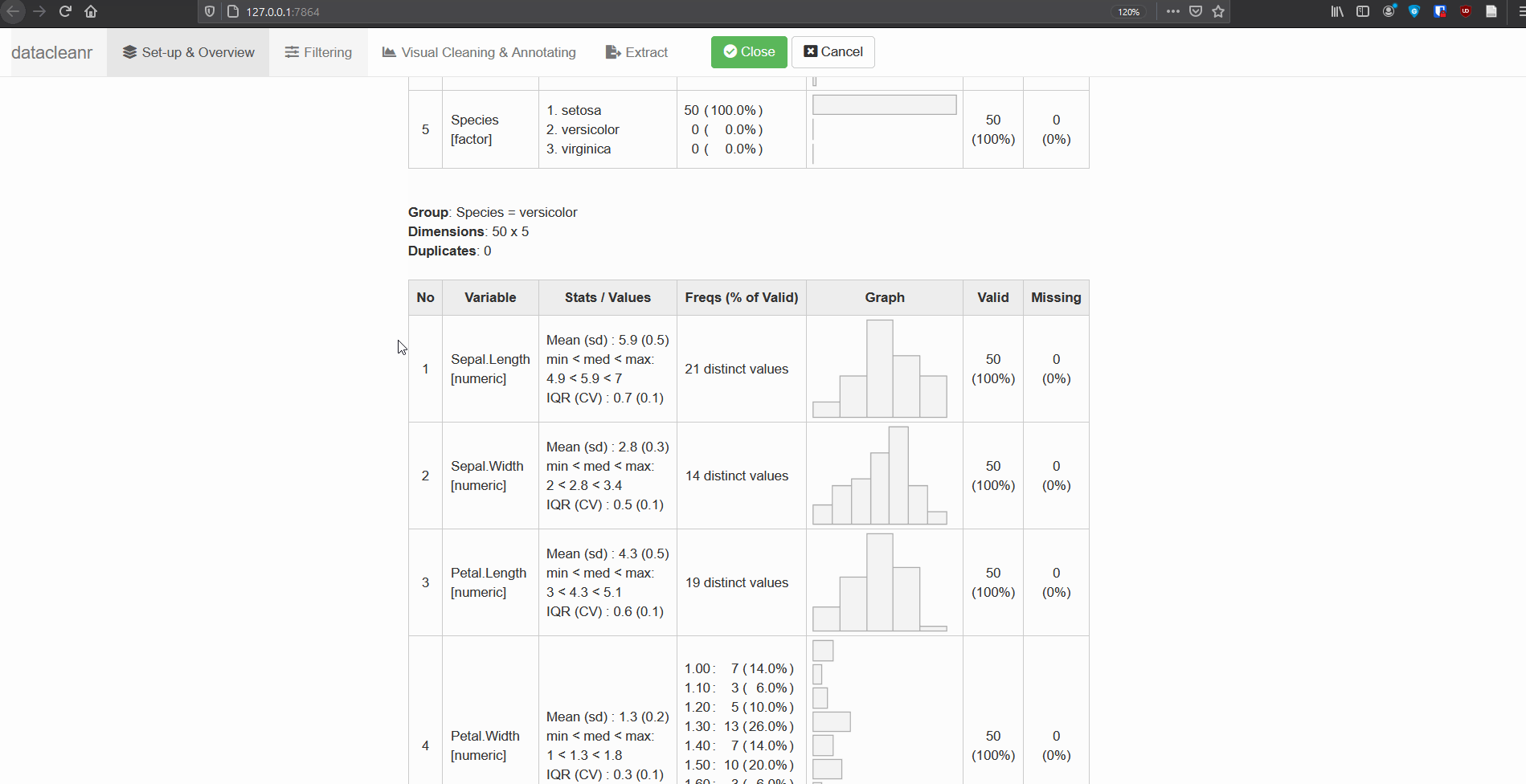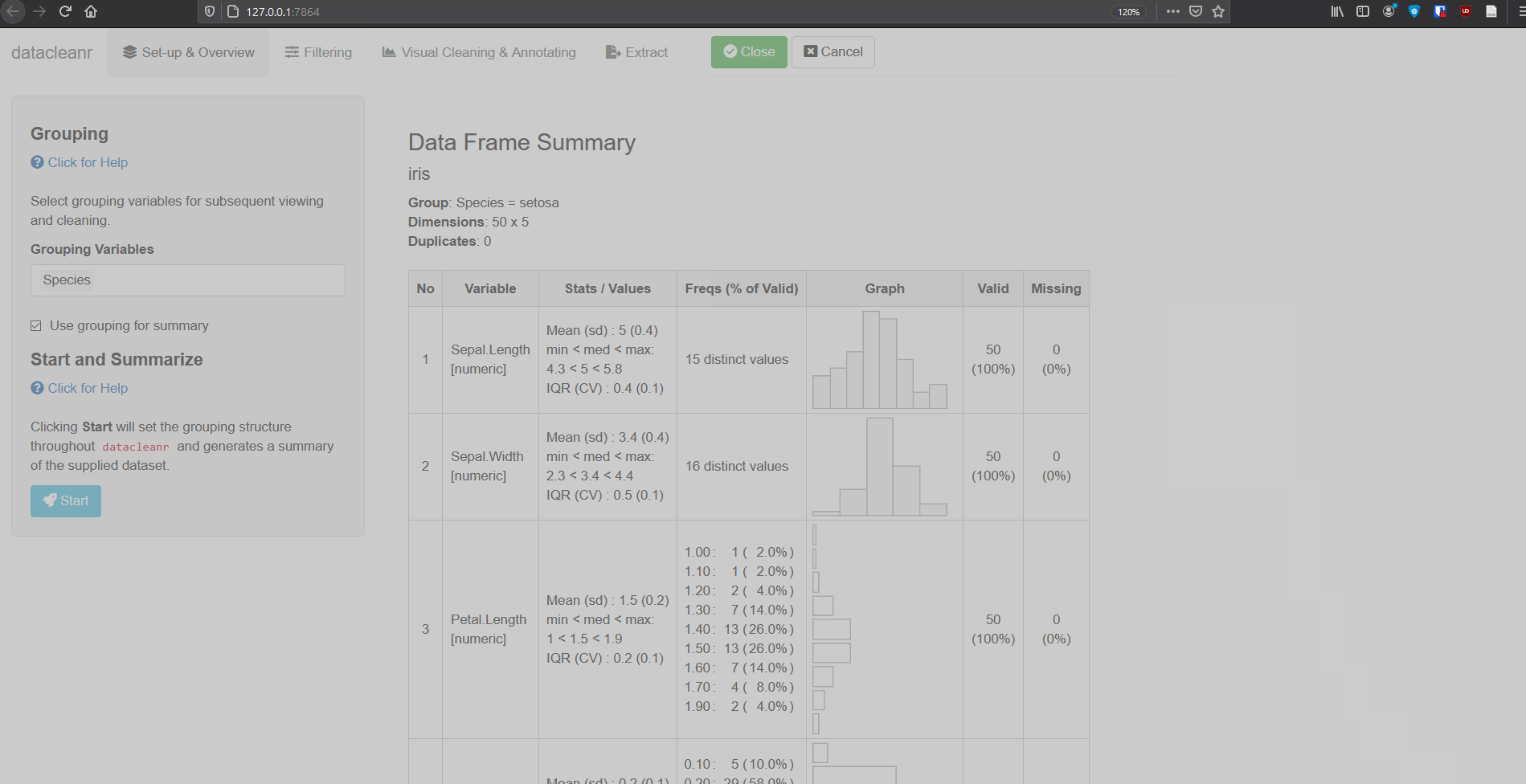
1. Set-up & Overview
Define the grouping structure (used throughout app for scoping filters and plotting), and generate an informative overview.
library(datacleanr)
# group by species
dcr_app(iris)
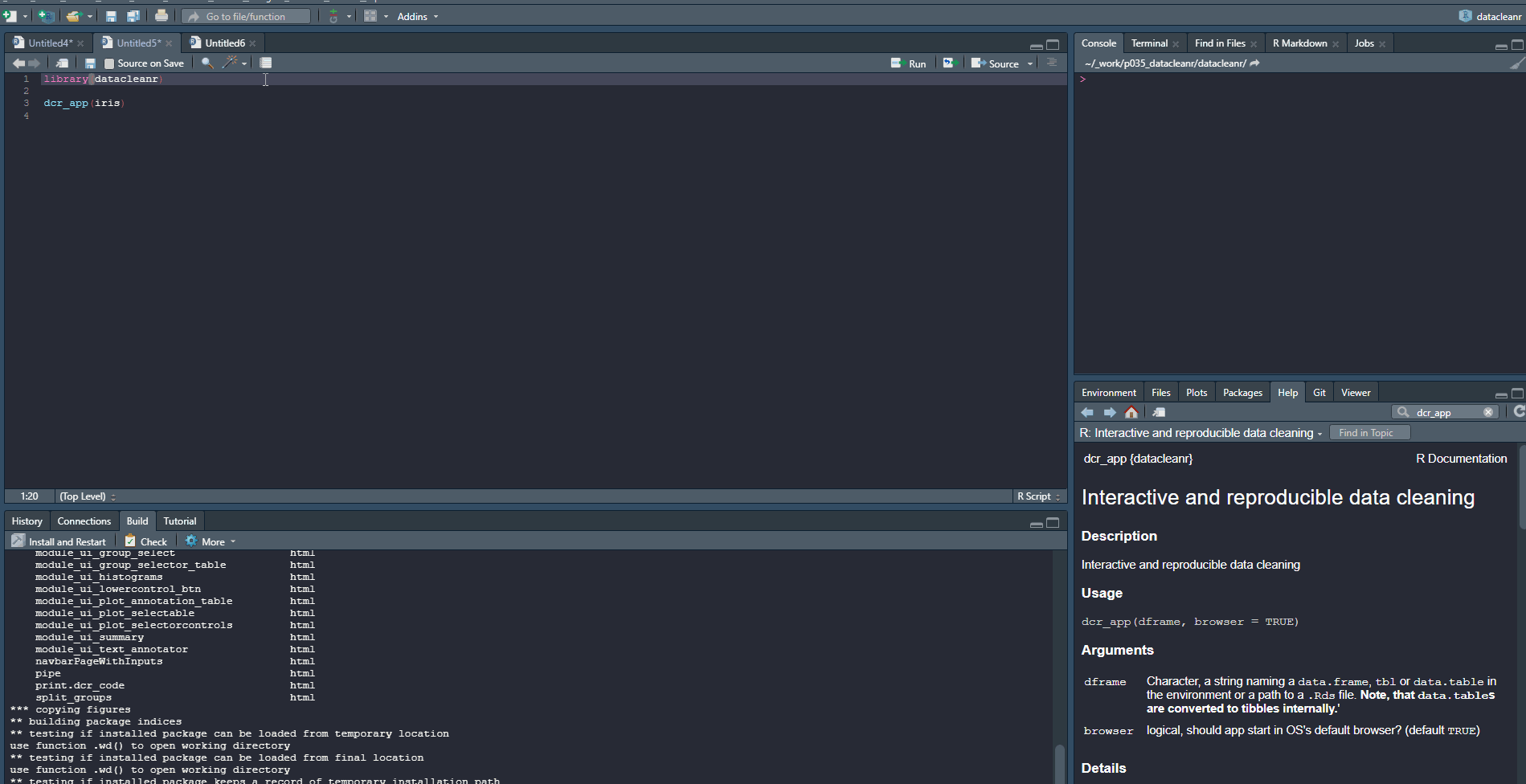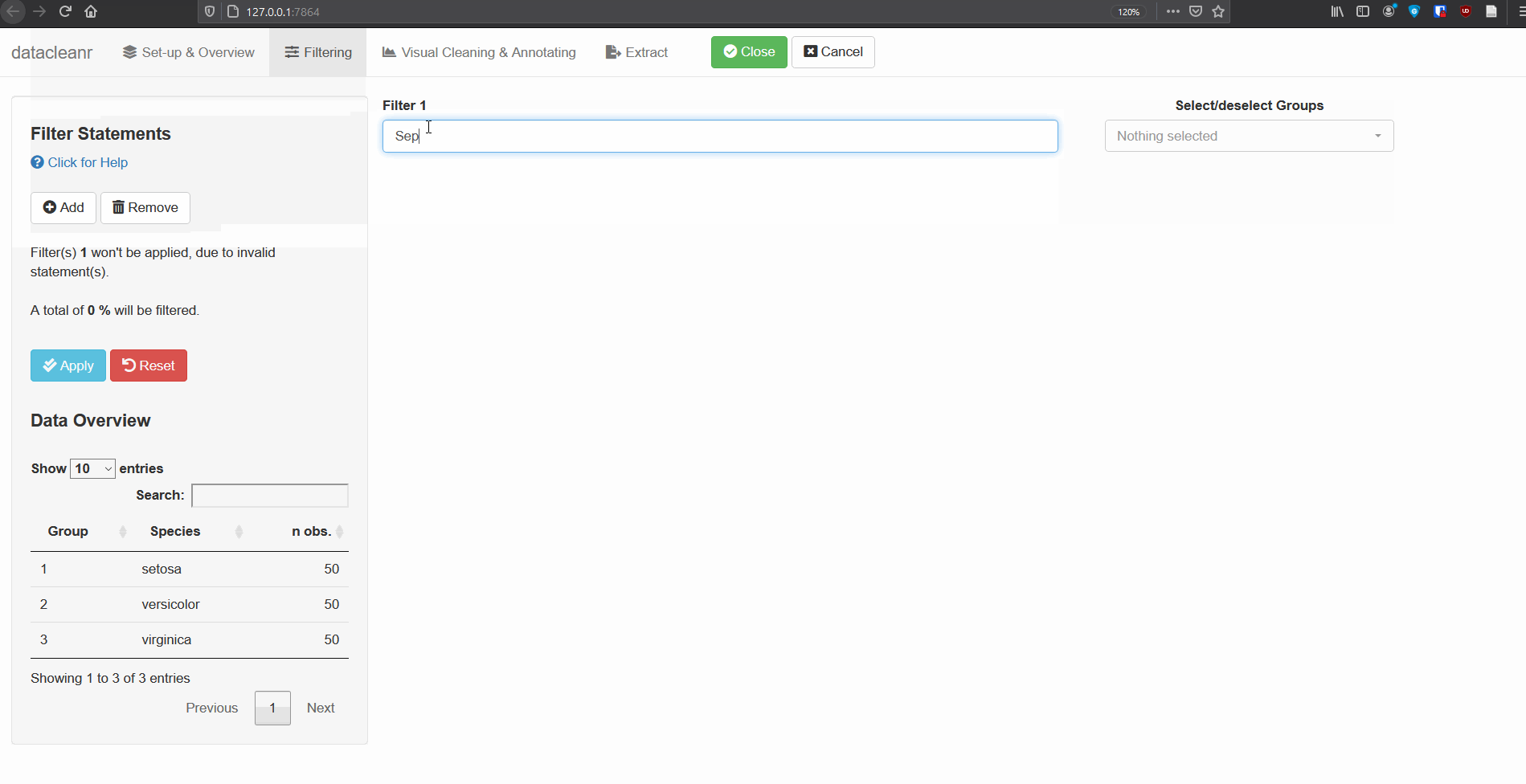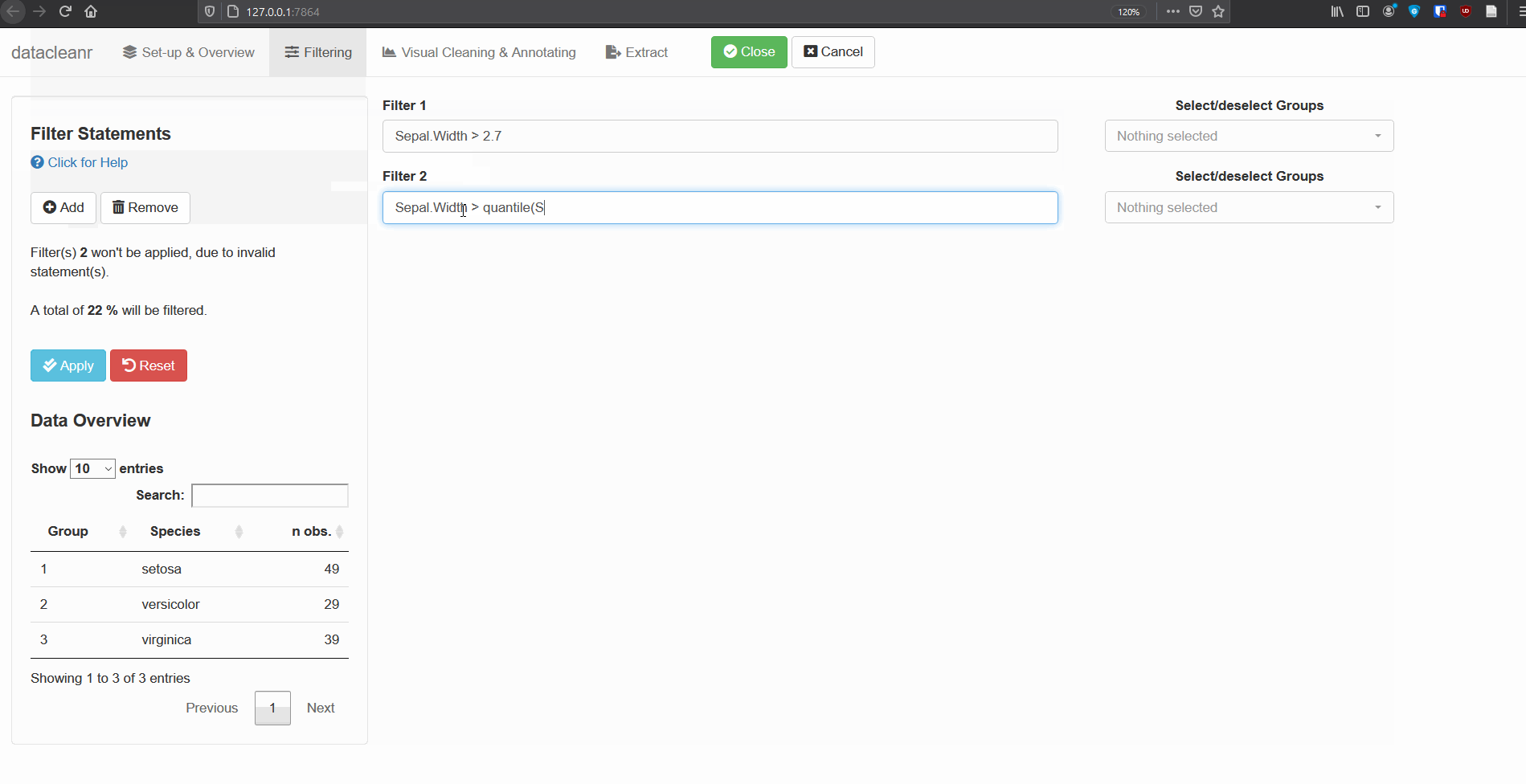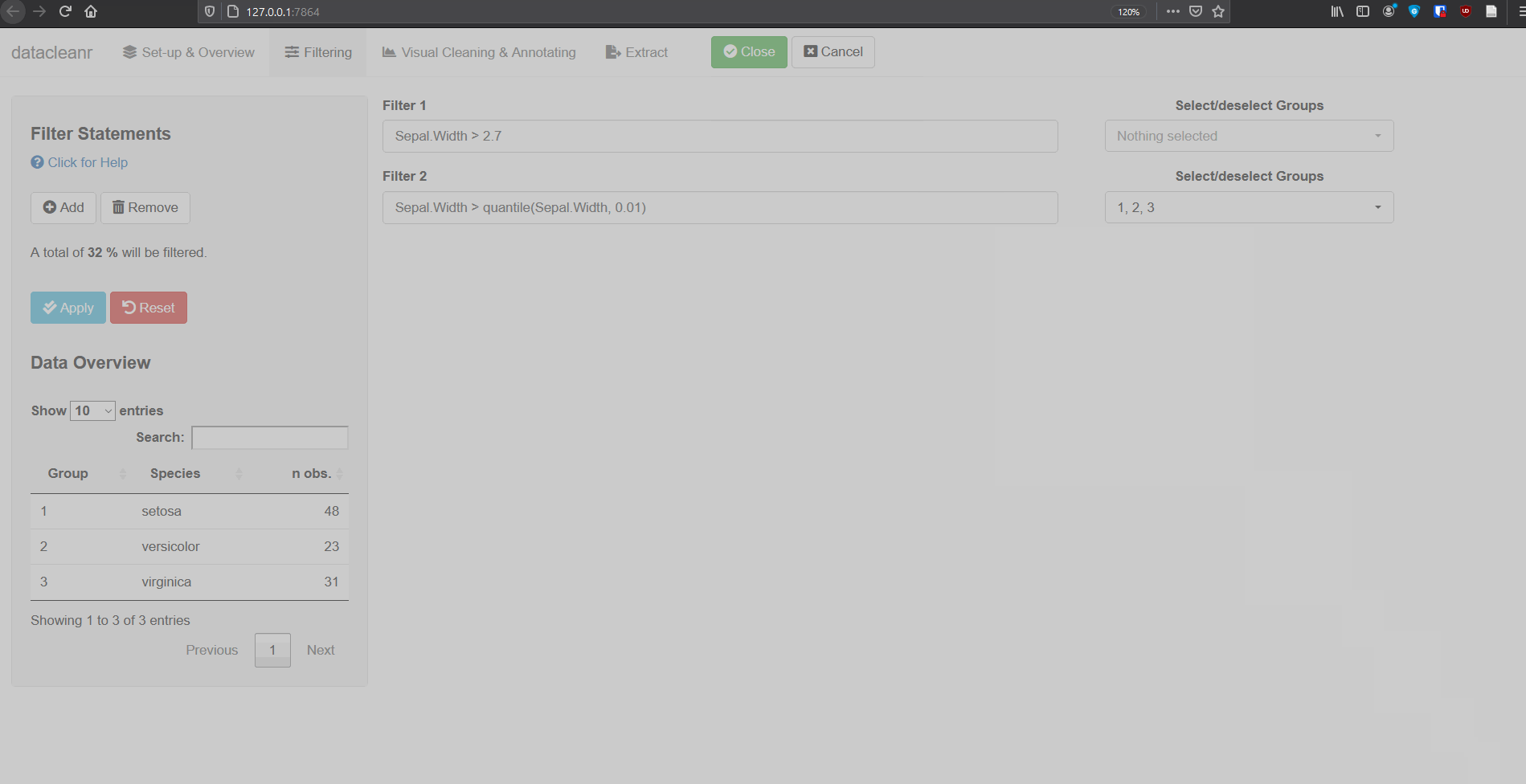
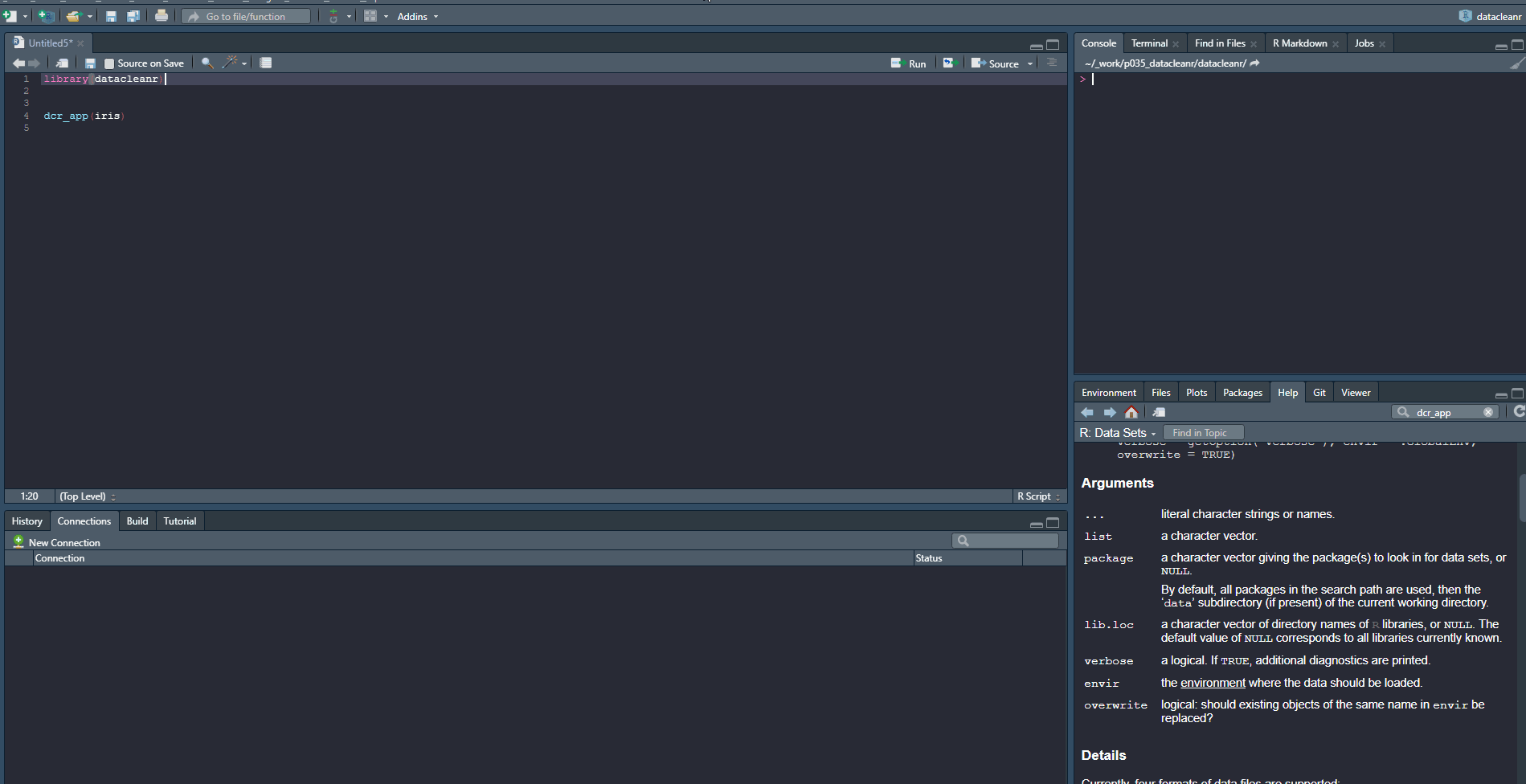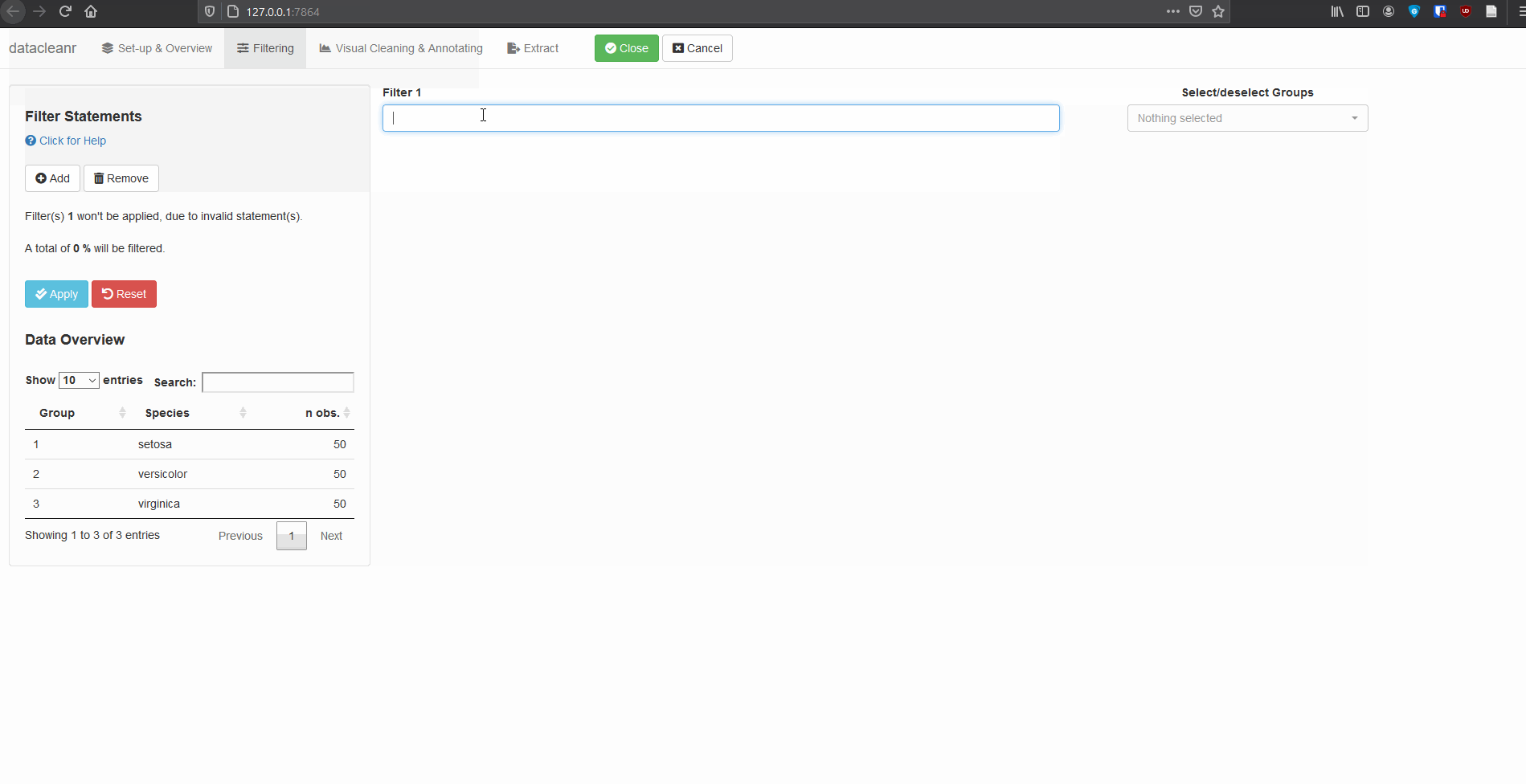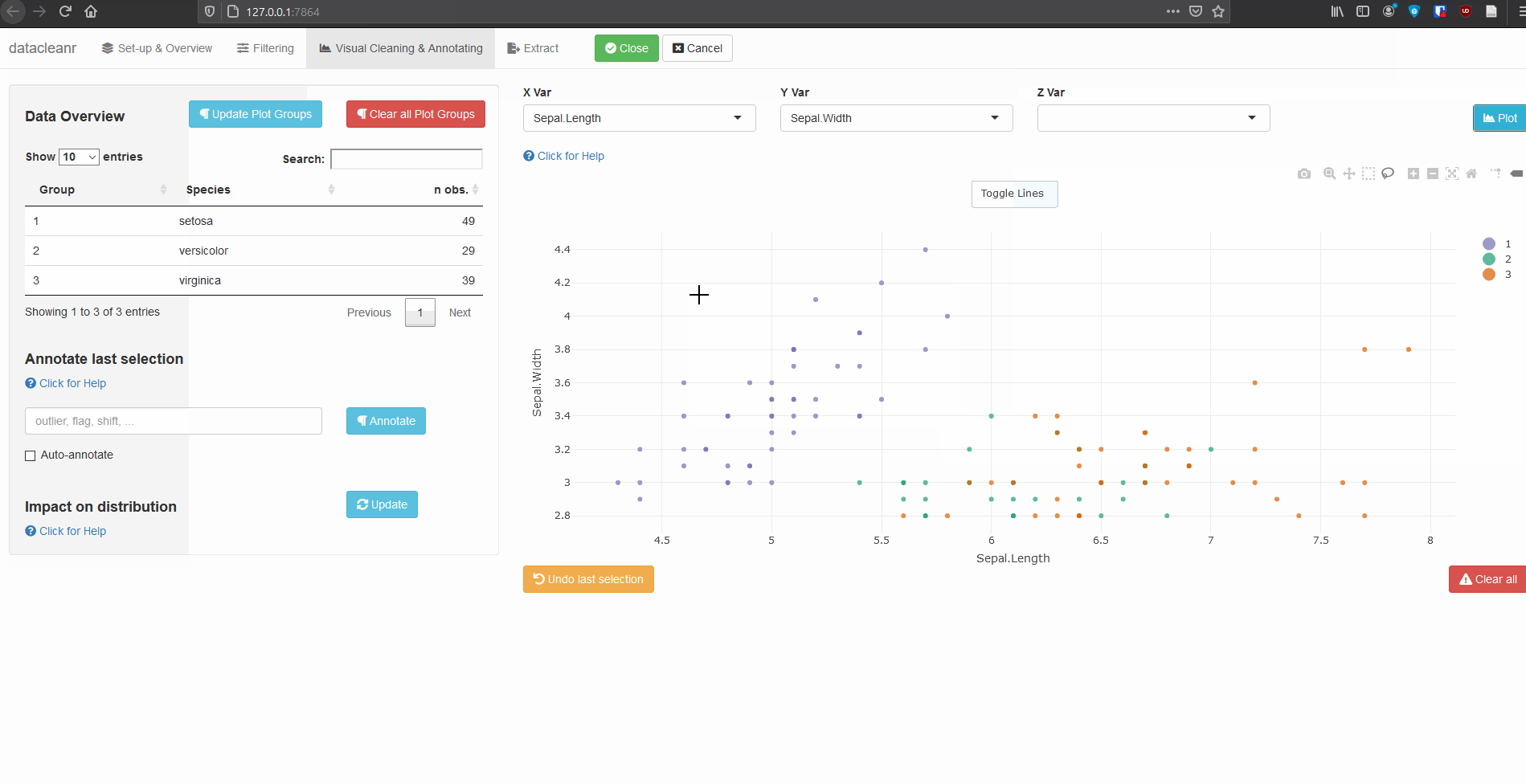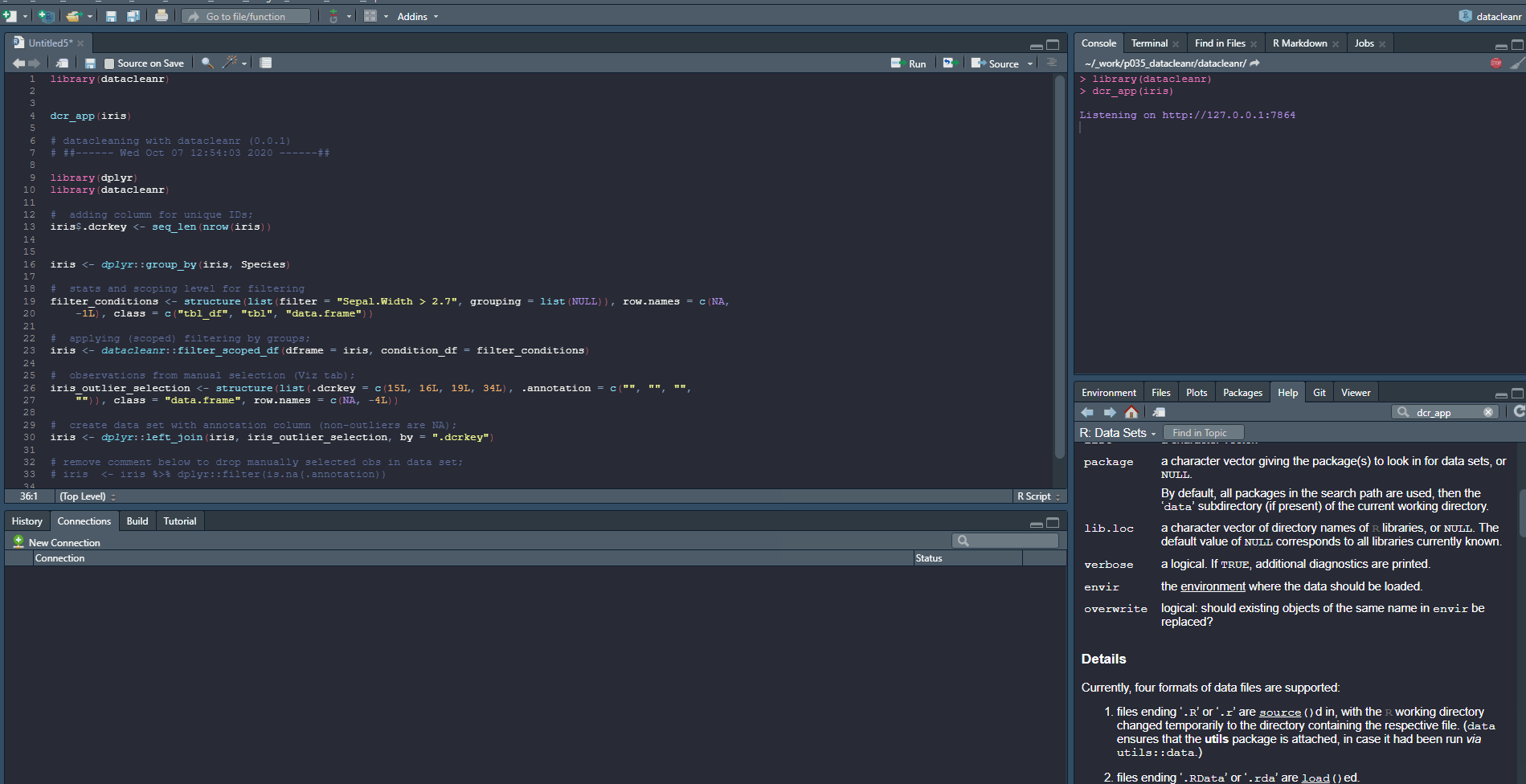
2. Filtering
Add/Remove filter statement boxes, and apply (valid) expressions - either to the entire data set, or scoped to individual groups. Filtering relies on R expressions passed to dplyr::filter(), so, for example, valid statements for iris are:
Species == 'setosa'
Species %in% c('setosa','versicolor')
Sepal.Width > quantile(Sepal.Width, 0.05)
Any function returning a logical vector (i.e. TRUE/FALSE), can be employed here!
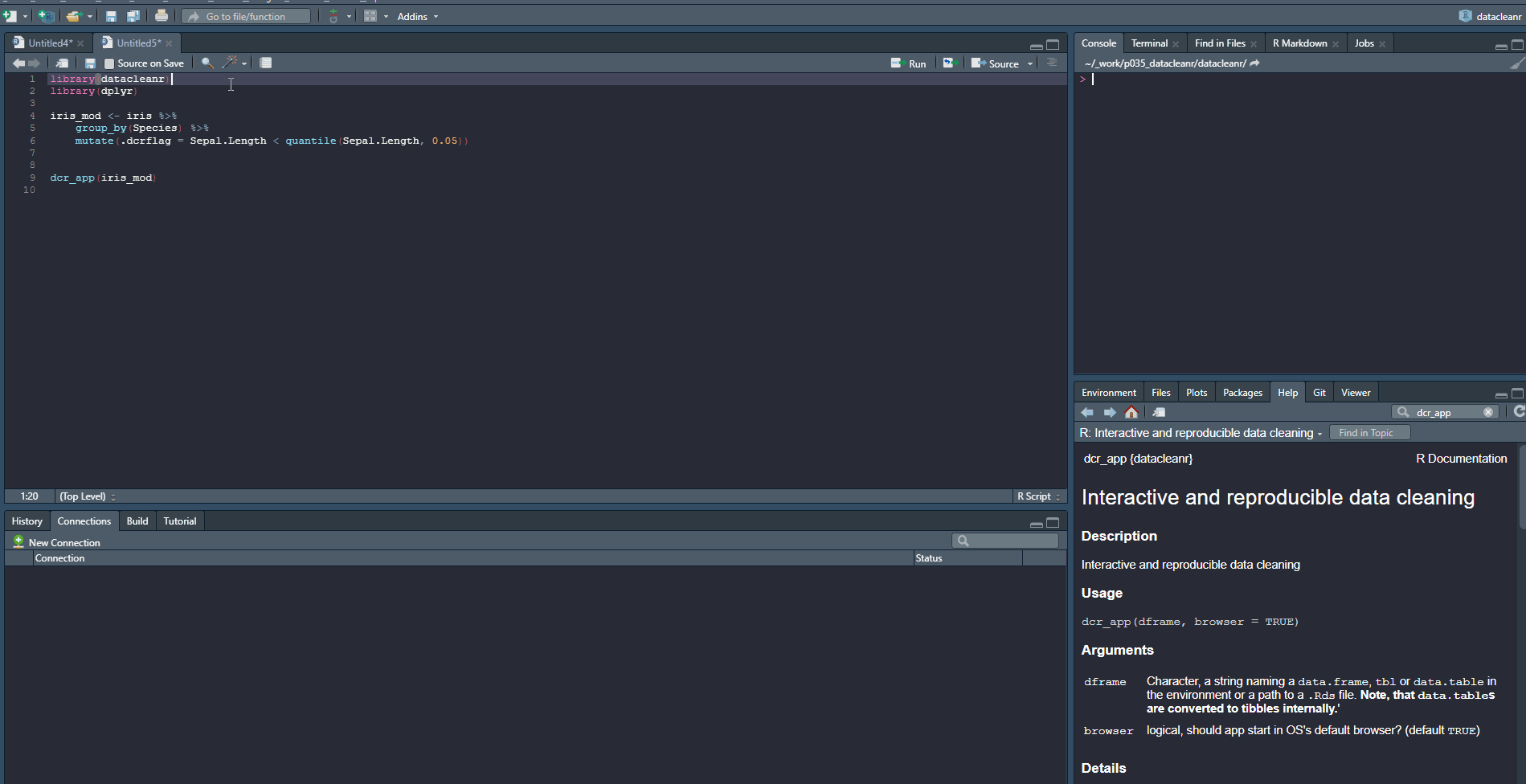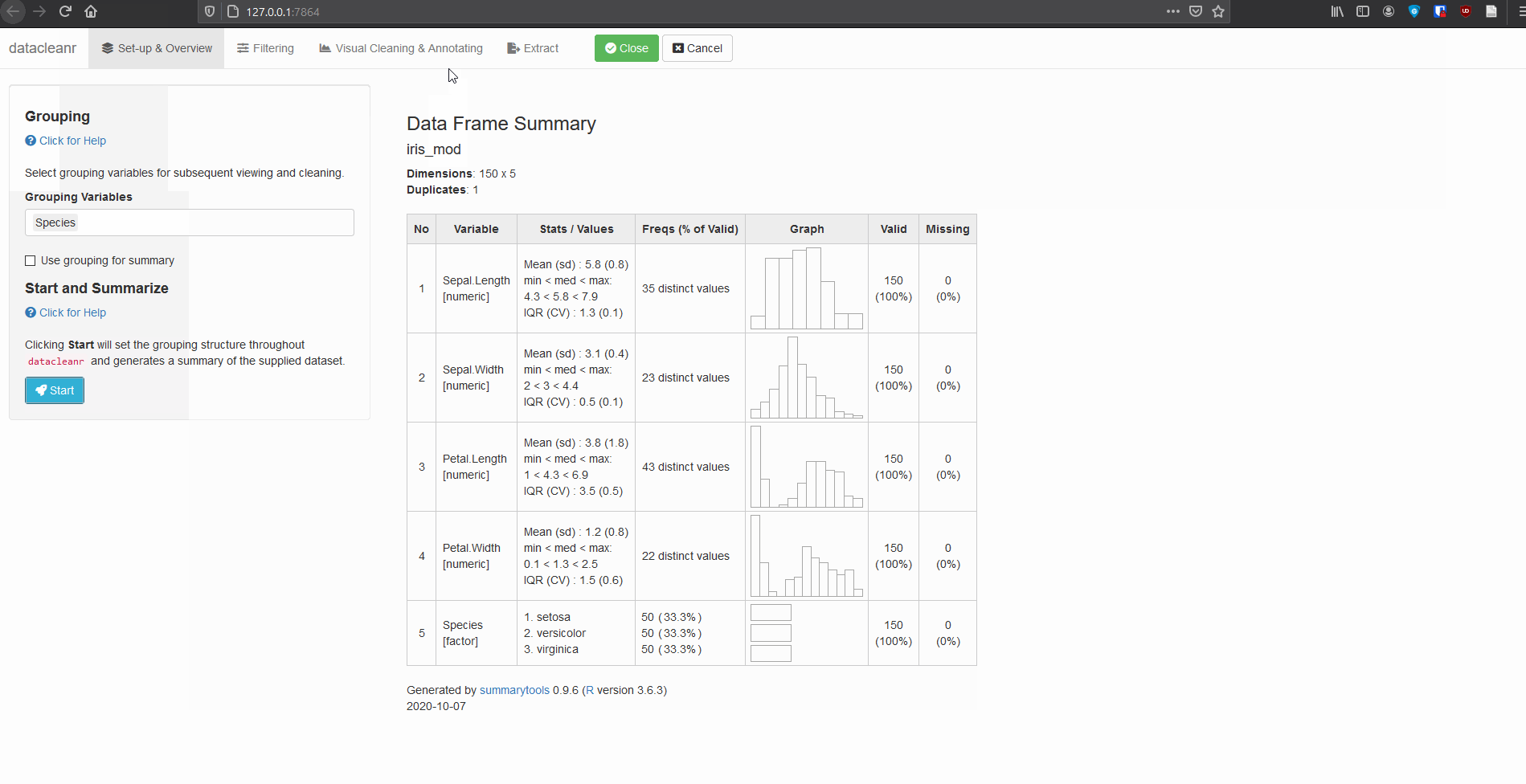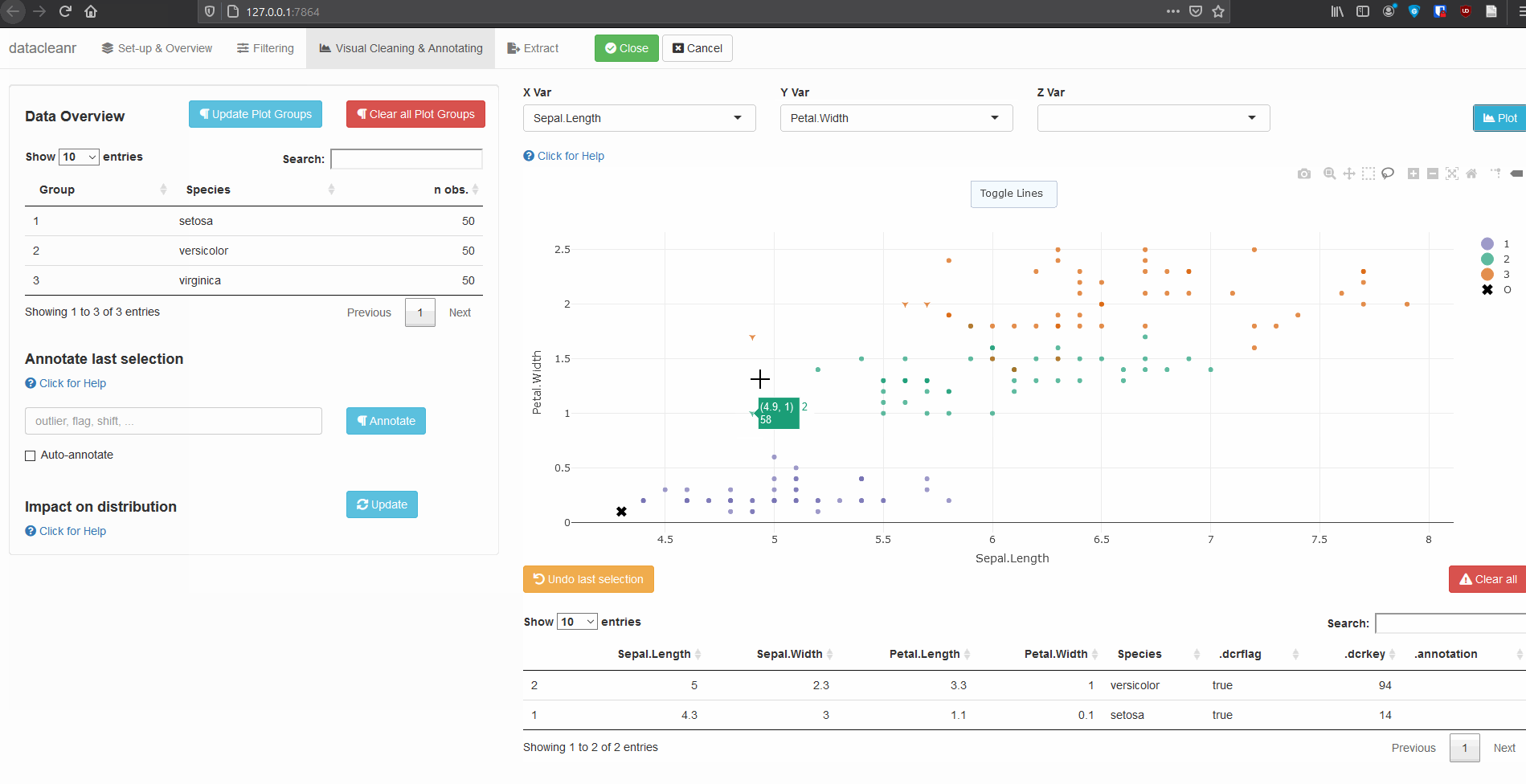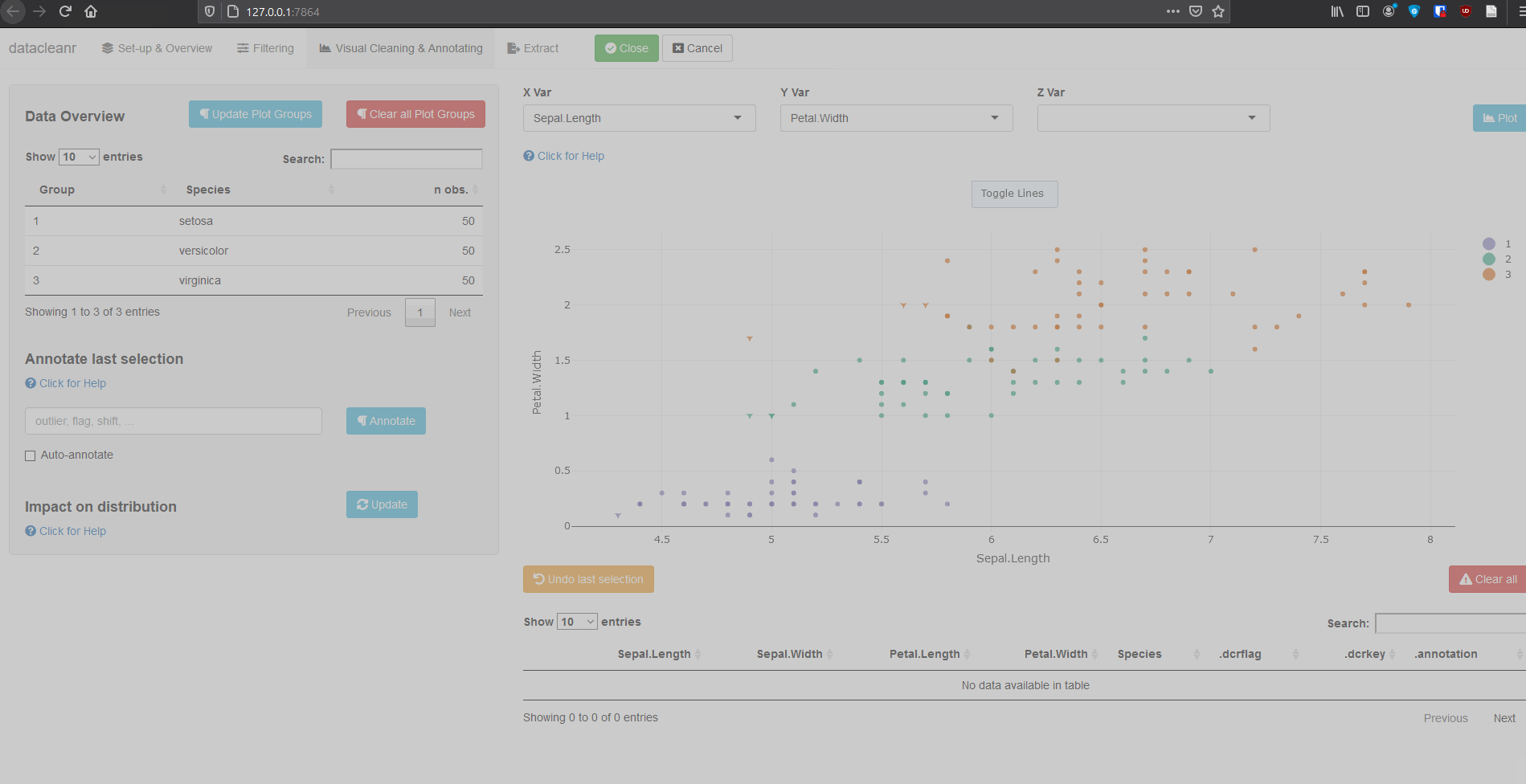
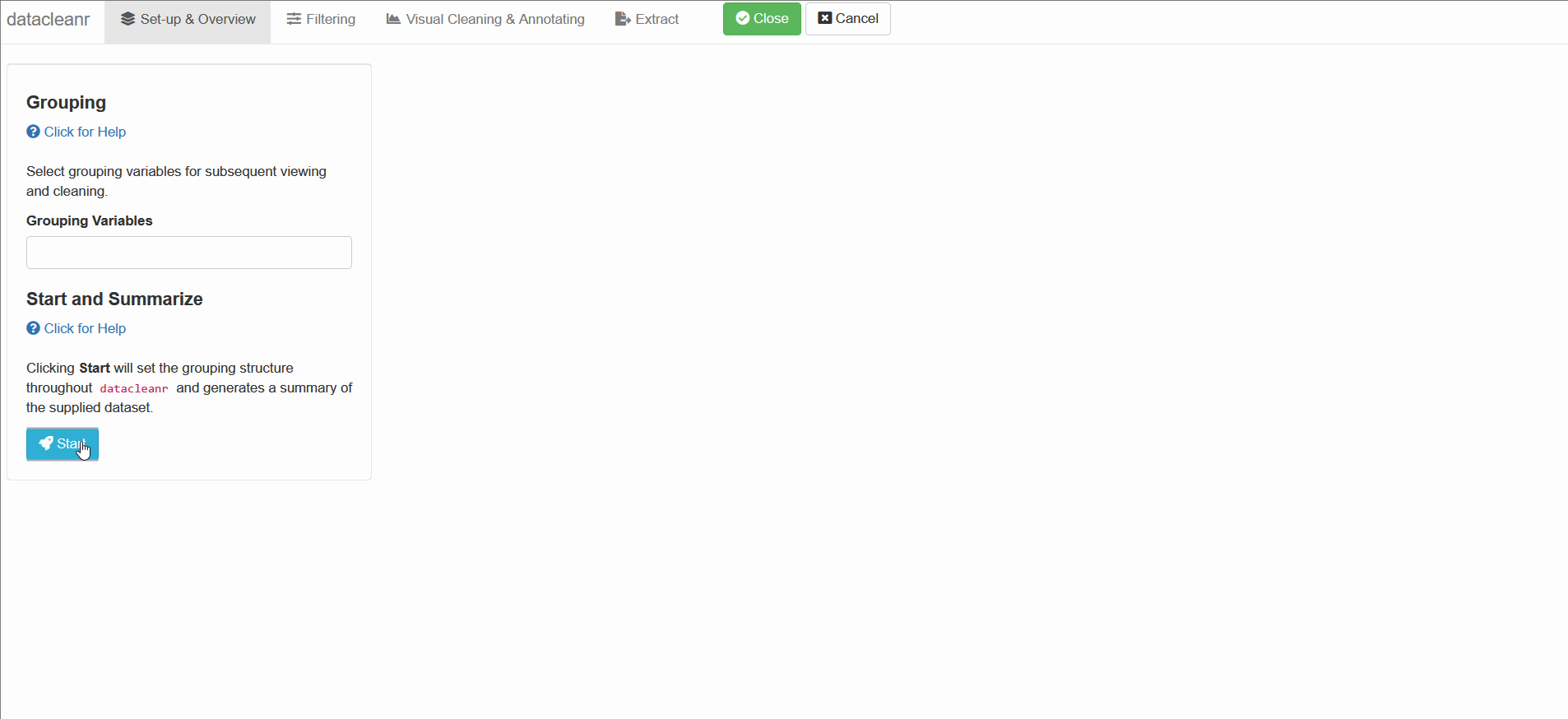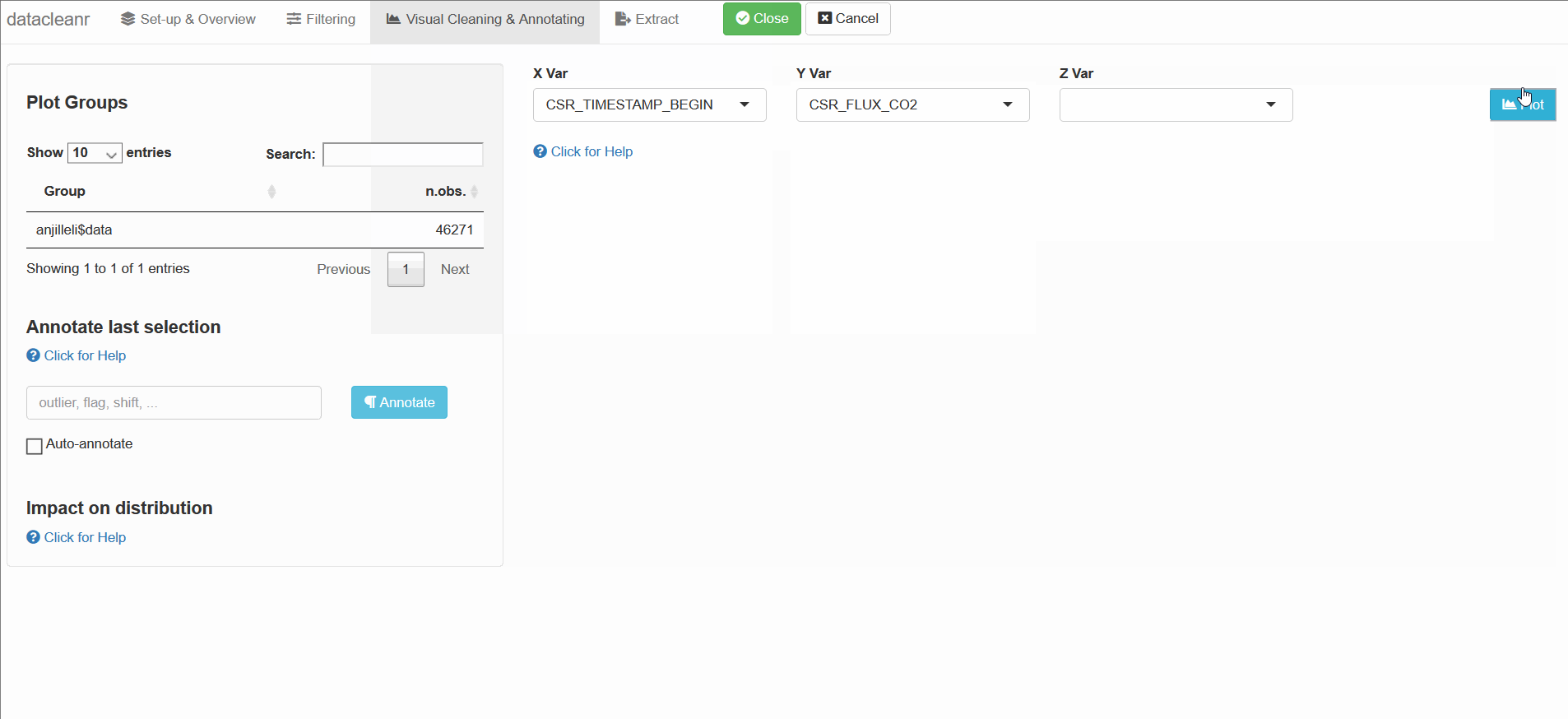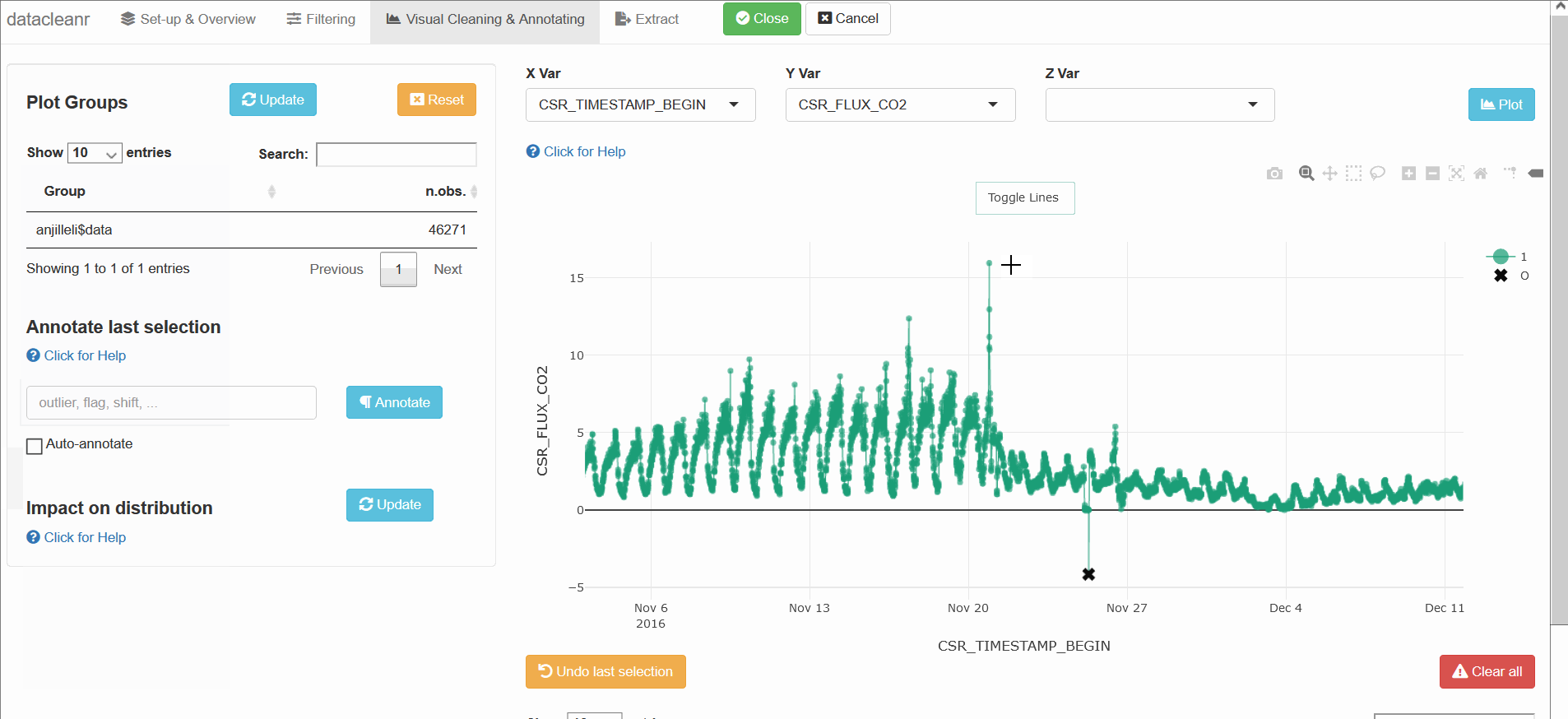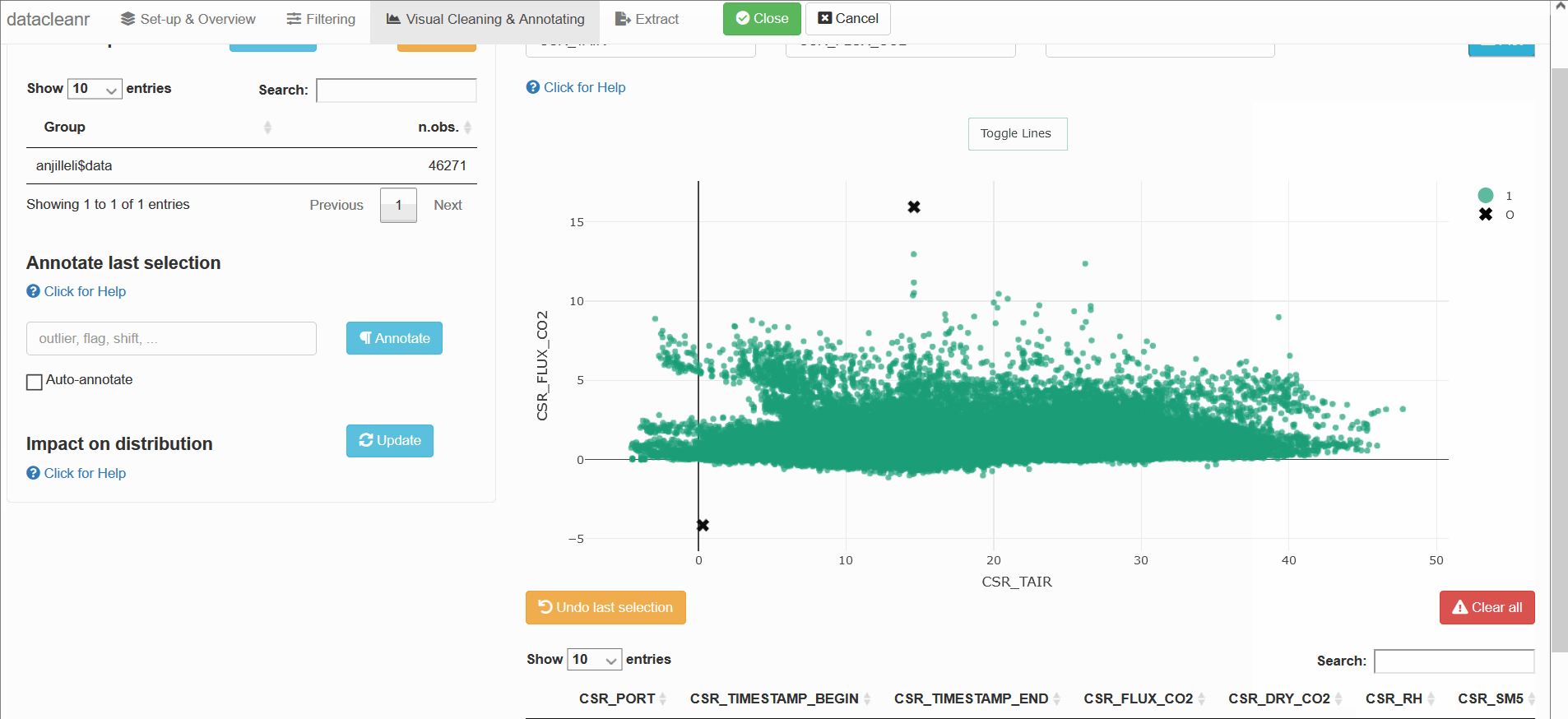
3. Visualizing and annotating
Interactive visualization allow seamless scrolling, panning and zooming to select and annotate individual observations (or sections with lasso/box select tool). Show and hide groups using the group selection table (left) or the legend (right).
3.1 General highlighting and annotating
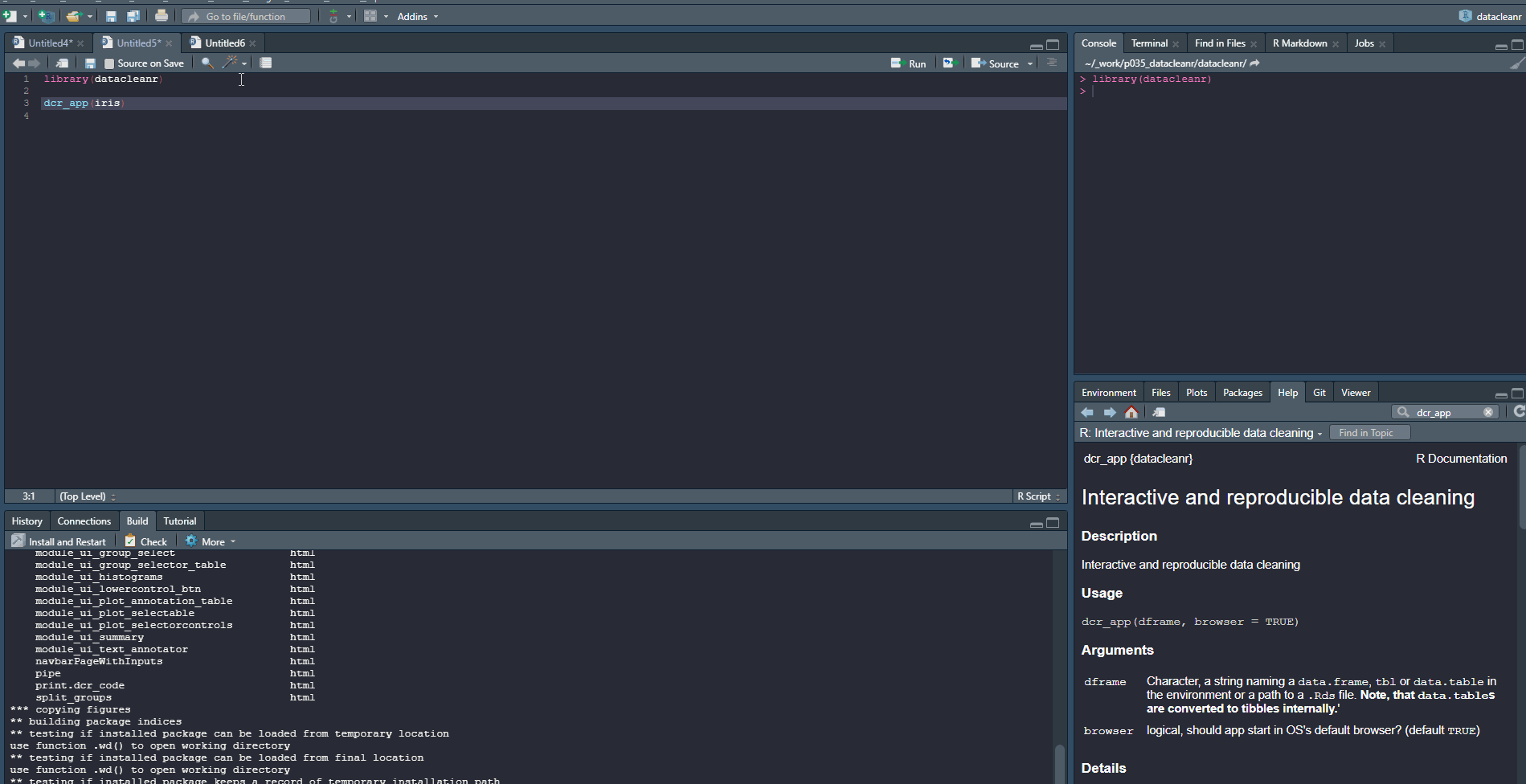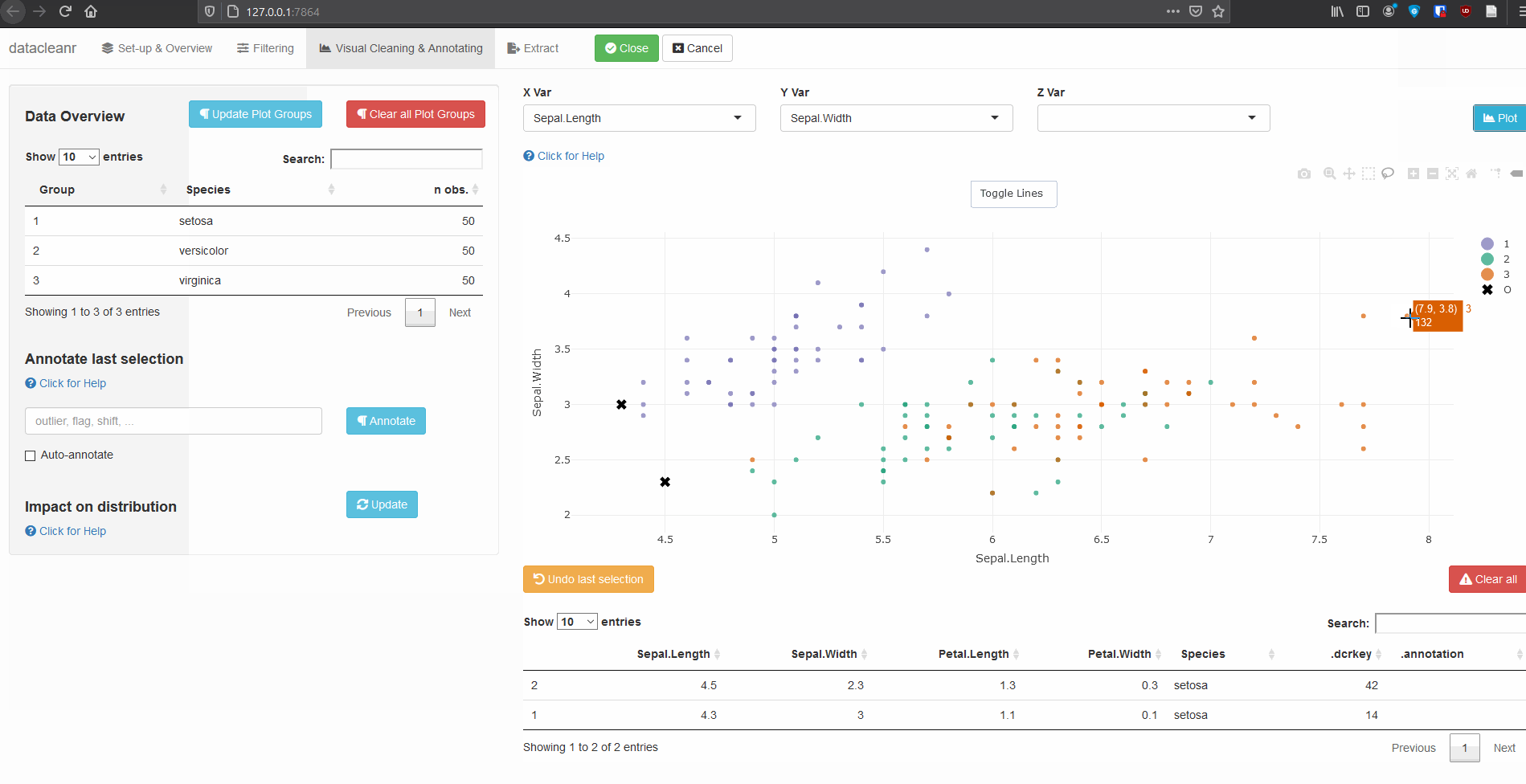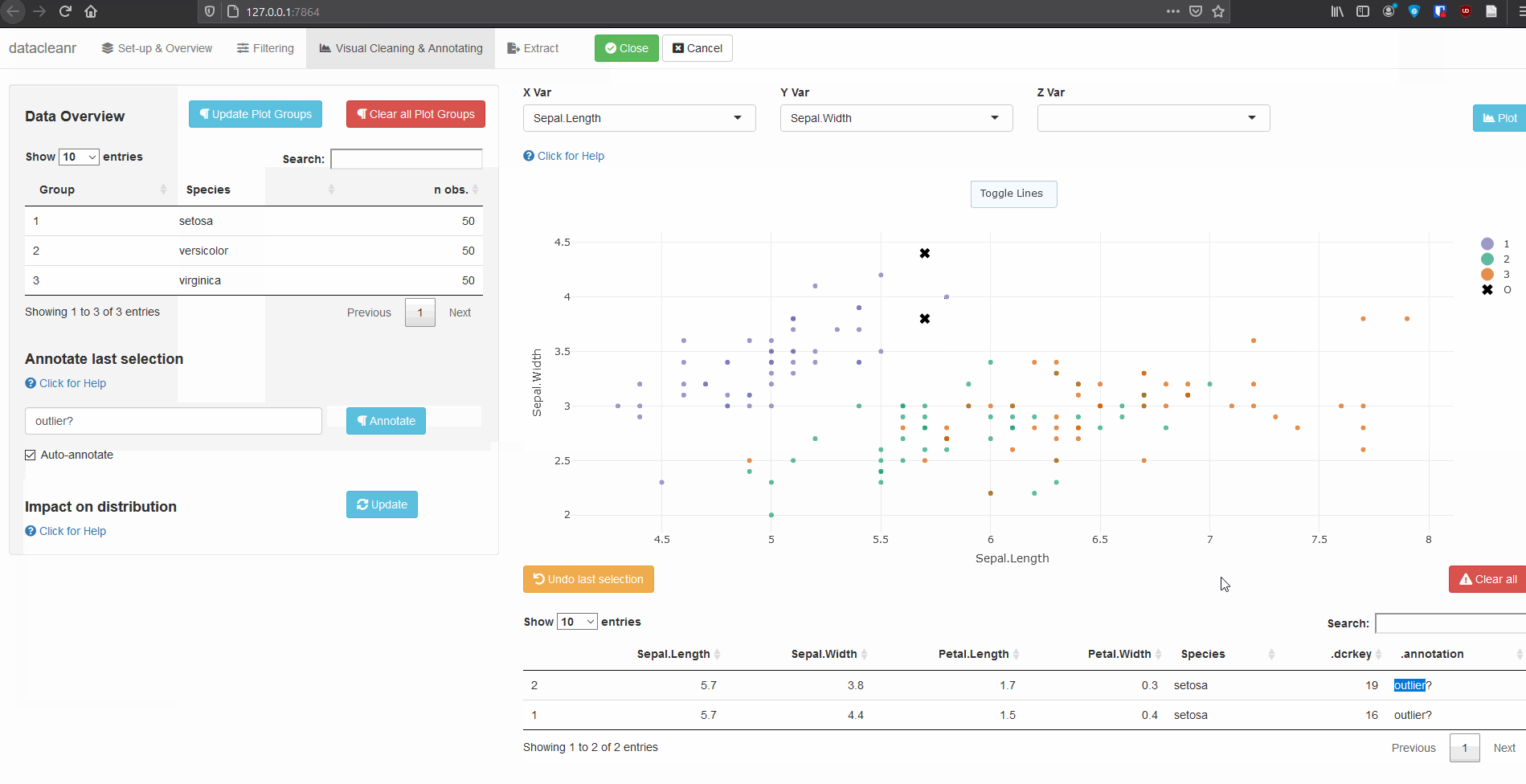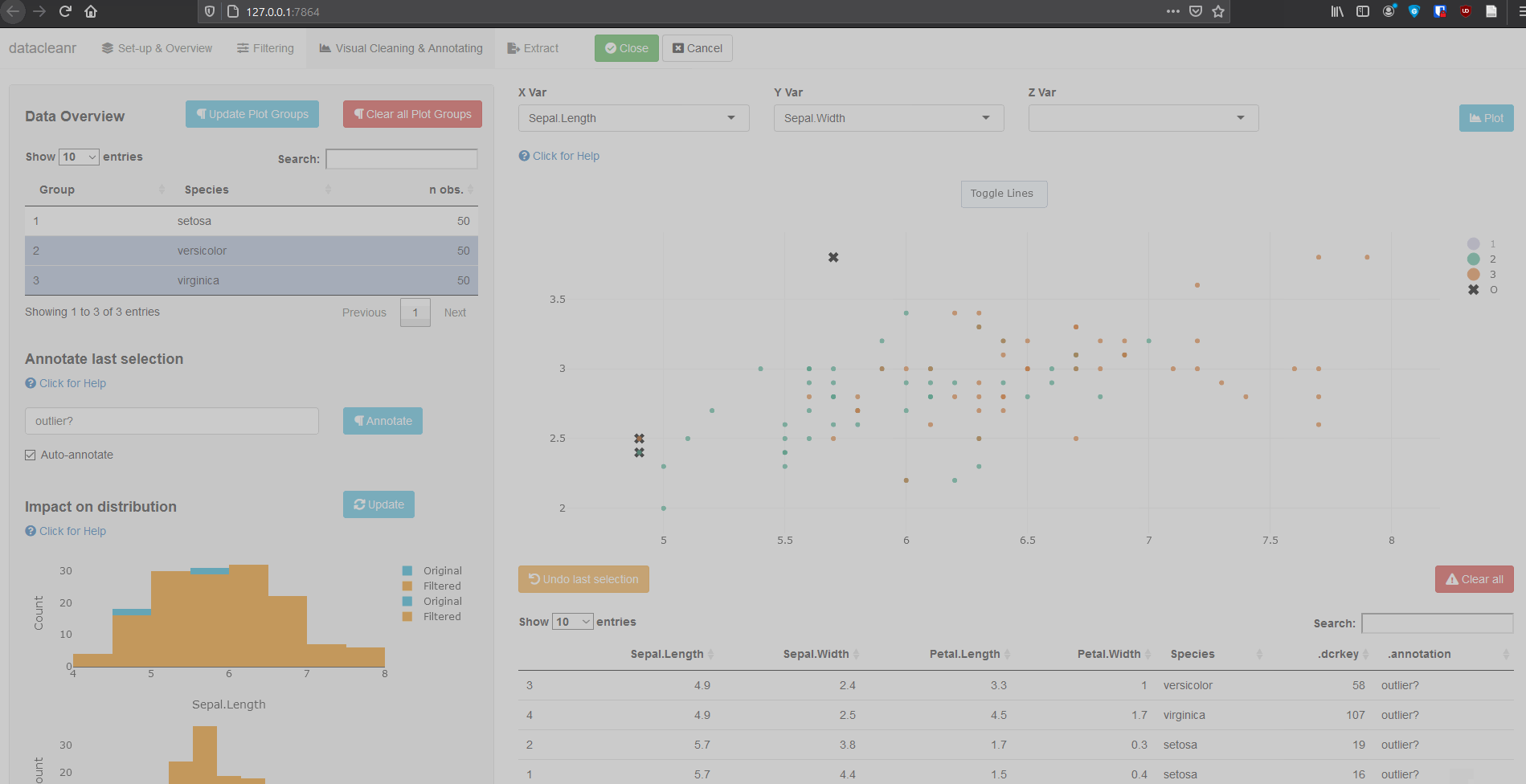
3.2 Using .dcrflag to interface with external QA/QC
library(datacleanr)
library(dplyr)
iris_mod <- iris %>%
group_by(Species) %>%
# .dcrflag provides additional visual cue in visualization tab
# based on TRUE/FALSE
mutate(.dcrflag = Sepal.Width < quantile(Sepal.Width, 0.05))
dcr_app(iris_mod)
3.3 Time Series
Any numeric or POSIXct column (in X or Y dimension) can be used to visualize time series. Use the Toggle Lines button above the plot to facilitate exploration.
Example 1:
library(dplyr)
dplyr::glimpse(treering)
tree_df <- data.frame(year = -6000:1979,
val = treering)
# make synthetic data
tree_data <- list(tree_A = tree_df,
tree_B = tree_df %>%
mutate(val = val + rnorm(nrow(.), 0.5, 0.2)),
tree_C = tree_df %>%
mutate(val = val + rnorm(nrow(.), mean = -0.03, 0.1))) %>%
bind_rows(.id = "tree")
# group by tree and inspect
dcr_app(tree_data)
(Note, selections are arbitrary and for demonstration only)
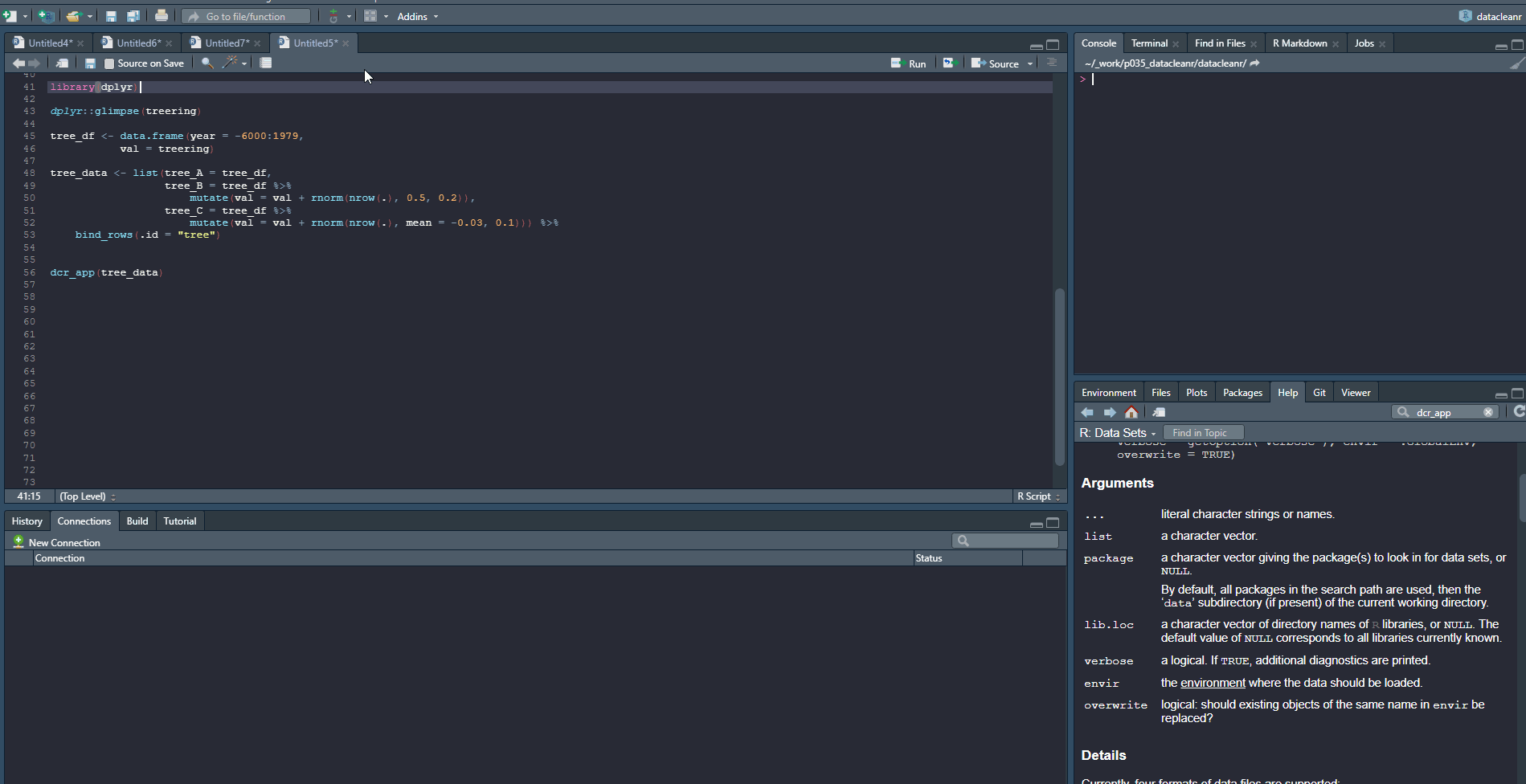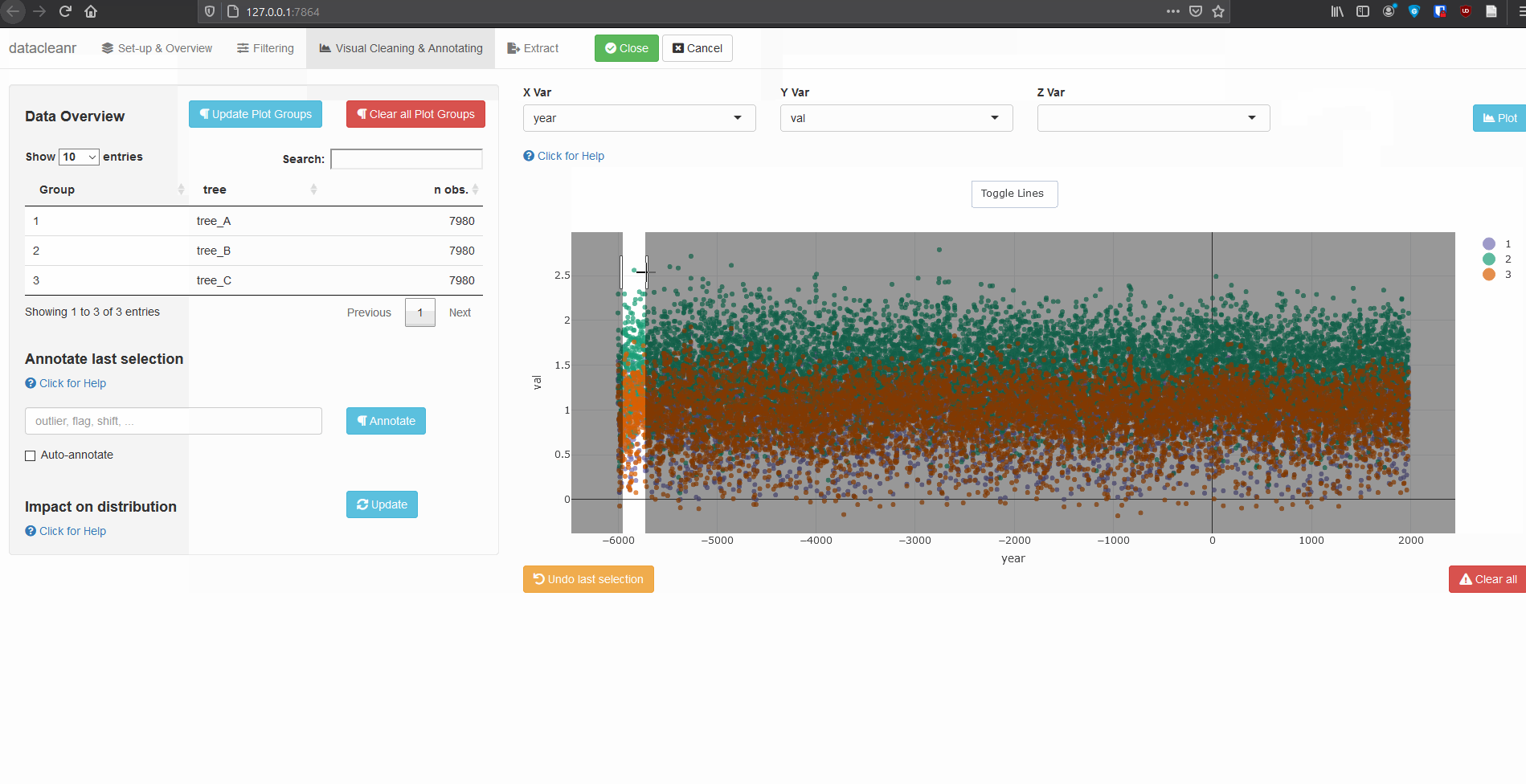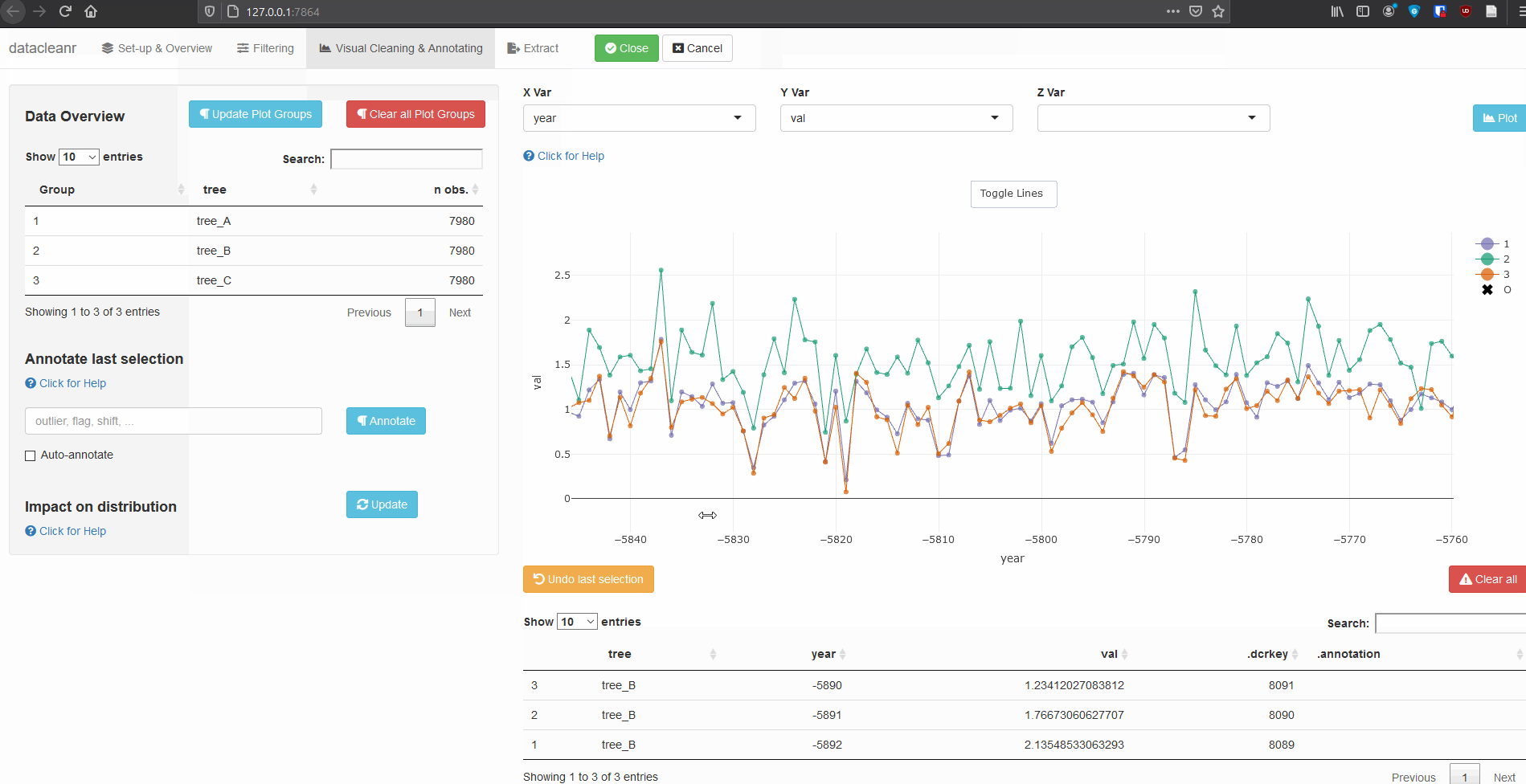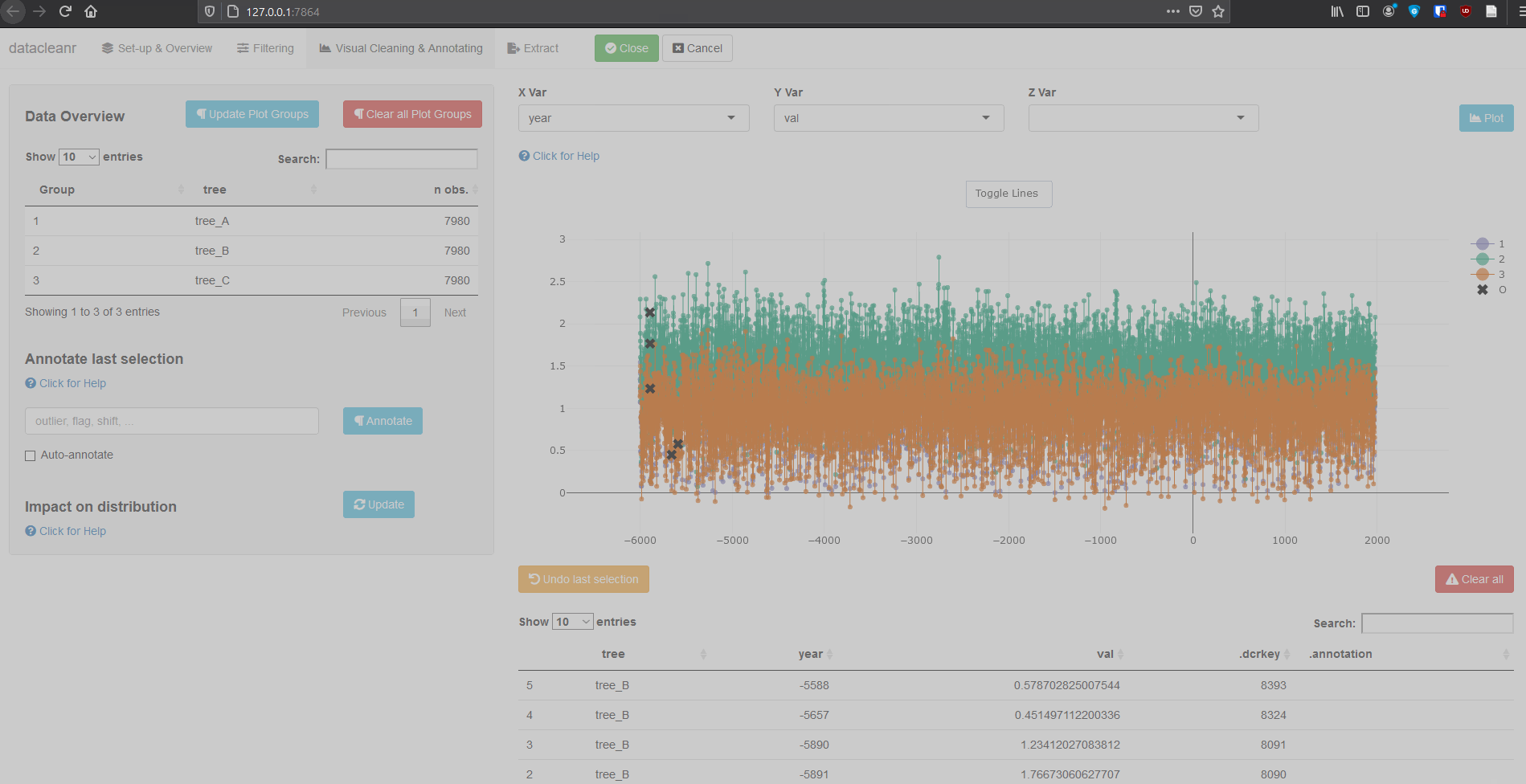
Example 2:
No GIF
library(dplyr)
library(lubridate)
data("storms", package = "dplyr")
storms_mod <- storms %>%
mutate(timestamp = lubridate::ymd_h(paste(year, month, day, hour)))
# Group by name (198 groups)
# Check "Emily"
dcr_app(storms_mod)
3.4 Spatial
Interactive maps rely on Mapbox for plotting. Therefore, you will need to make an account, from which an access token needs to be copied into your .Renviron (e.g. MAPBOX_TOKEN=your_copied_token). A simple way to do this is using the convenient usethis package to access the file:
usethis::edit_r_environ()
Select columns lon and lat for plotting to get started.
Example 1
library(datacleanr)
library(dplyr)
airport_data <- read.csv('https://plotly-r.com/data-raw/airport_locations.csv') %>%
rename(lon = long)
# group by state
dcr_app(airport_data)
Example 2
No GIF
library(dplyr)
library(lubridate)
data("storms", package = "dplyr")
storms_mod <- storms %>%
rename(lon = long)
# Group by name (198 groups)
# Check "Bonnie"
dcr_app(storms_mod)
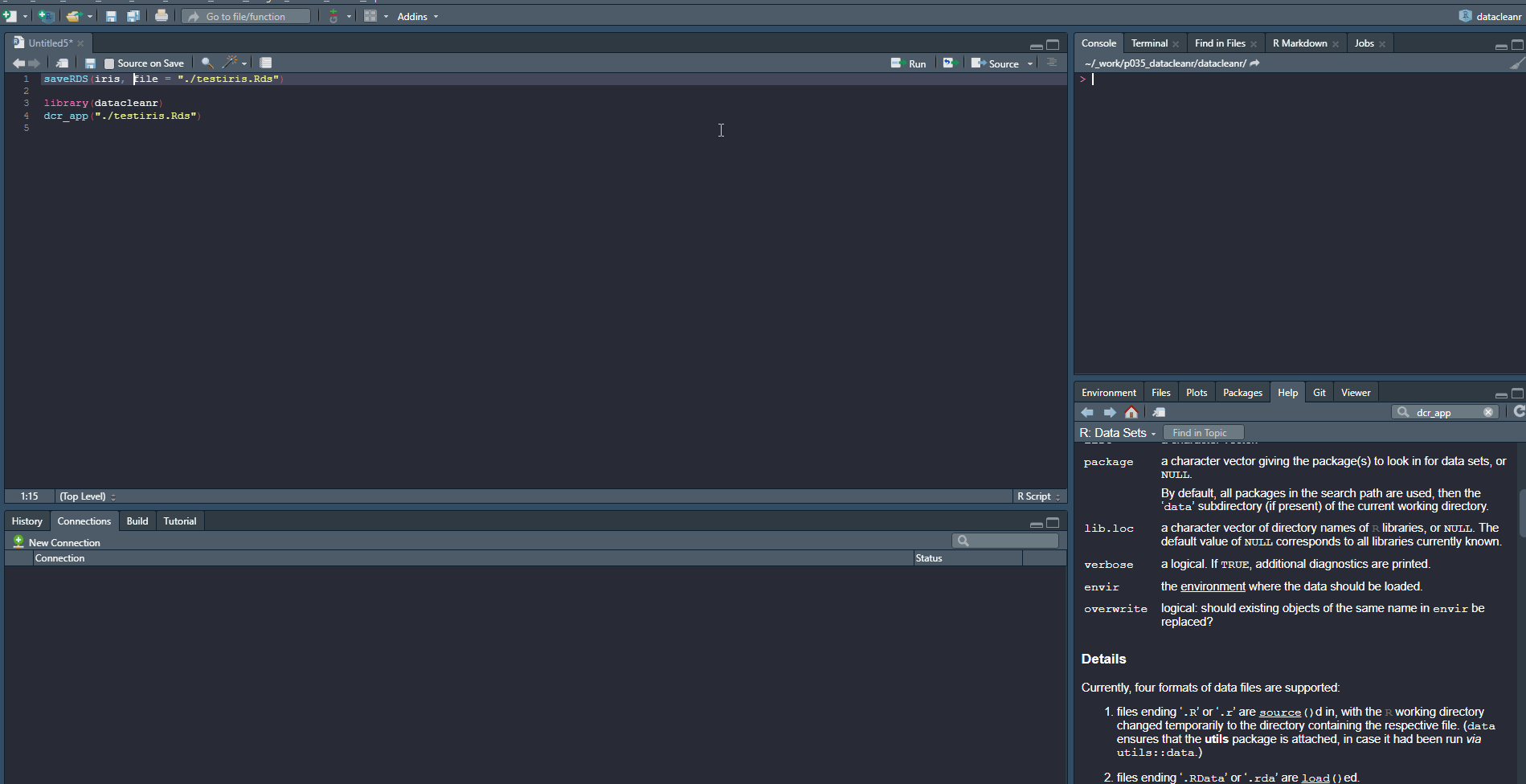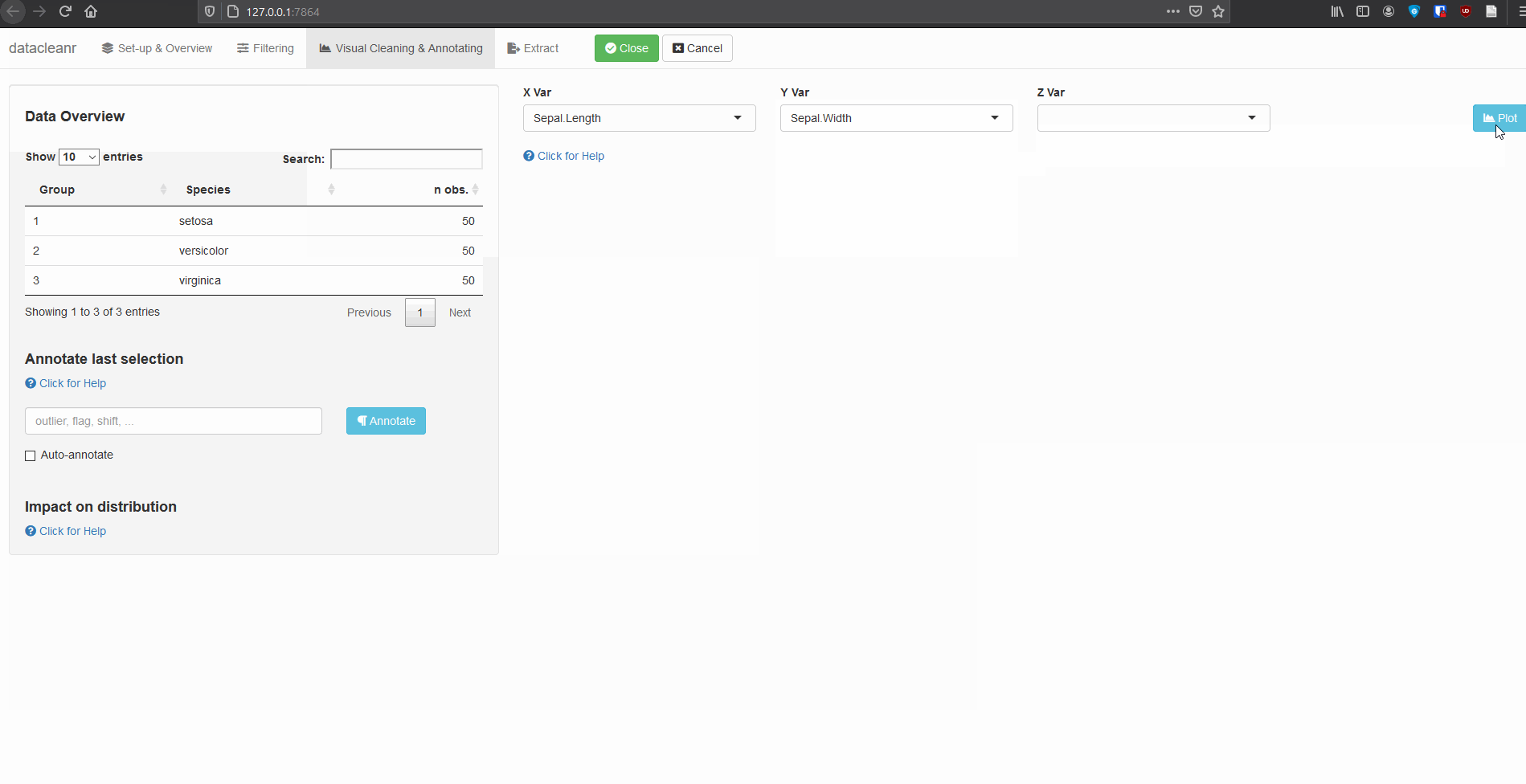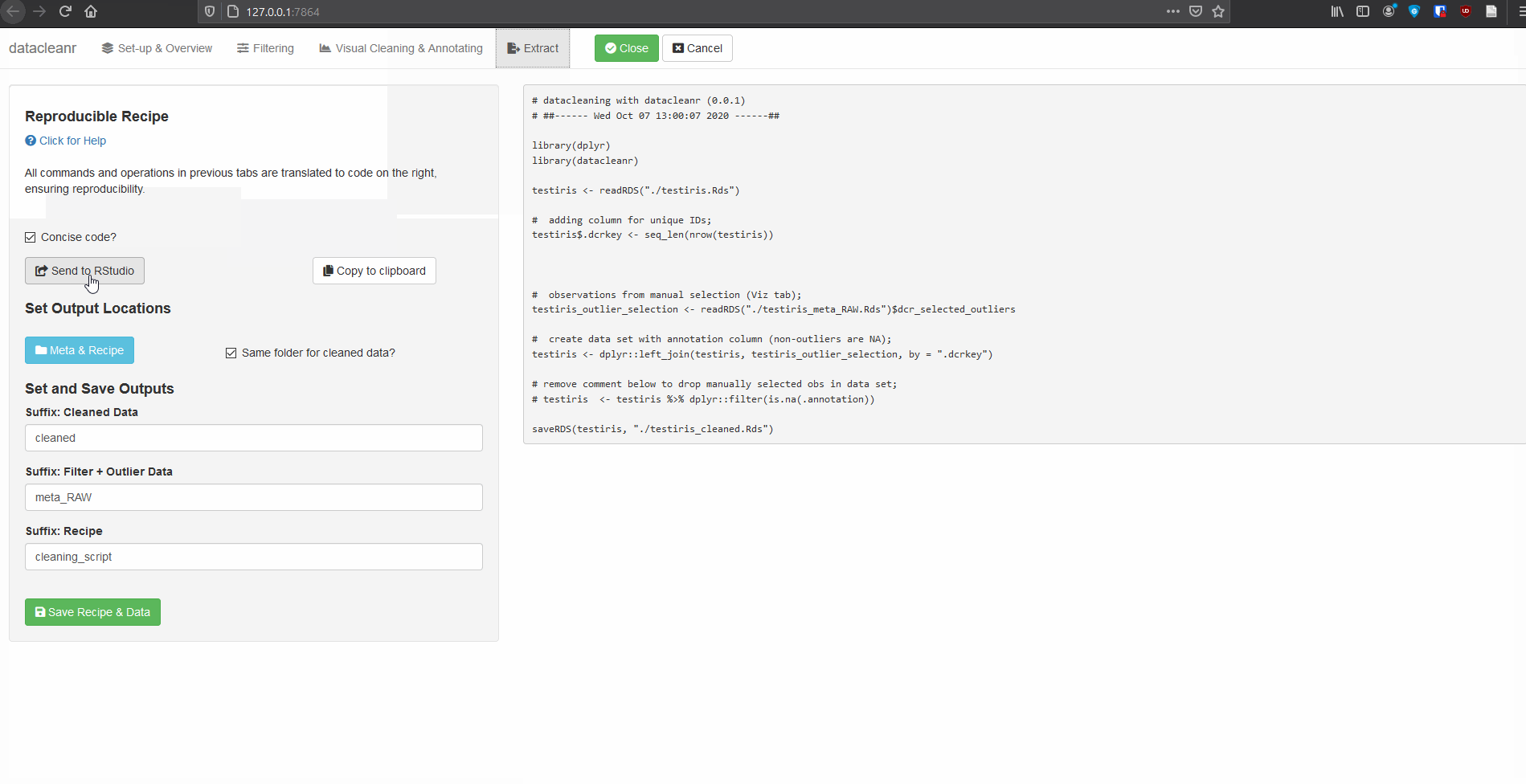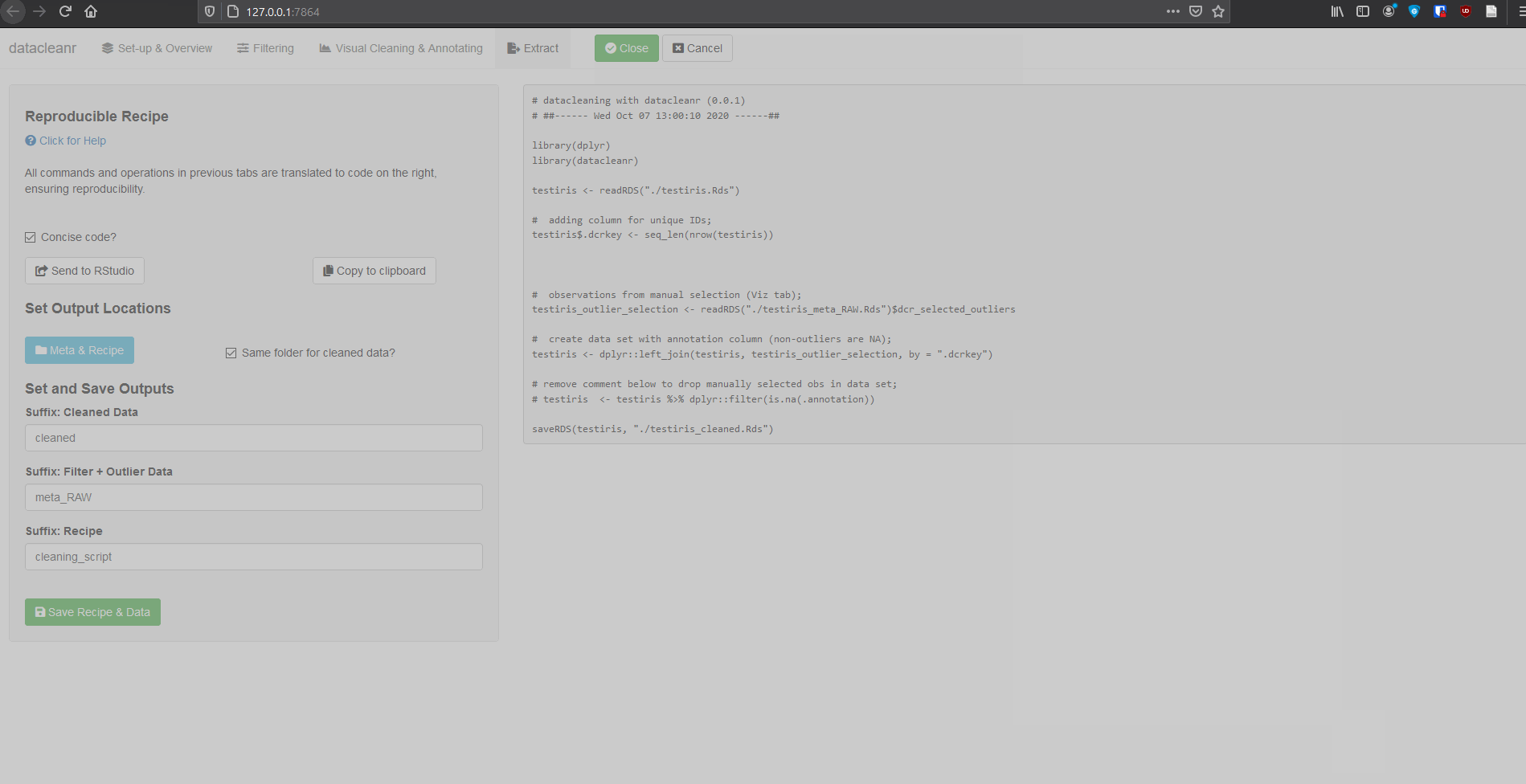
4. Extract (Reproducible Recipe)
All grouping, filtering and selections/annotations are translated to R code, which can be sent to an RStudio script, copied to the clipboard, or - when dcr_app is launched with a file path - save options are made available. For large selections/annotations we recommend saving the script separately, and sourcing it (i.e. source("your_datacleanr_script.R")) during later analyses.
Caution: When selections / annotations are greater than ~ 1000 points, it is recommended to use datacleanr with an *.RDS file (see below). This is because the resulting Reproducible Recipe (script) can slow down the RStudio IDE, if it has more than a few thousand lines.The next version of datacleanr will allow choosing between script-only recipes, and the option with an the intermediate file for storing annotations. Both approaches with their current implementation are shown shown below.
Example 1
Launching with an object from R:
library(datacleanr)
dcr_app(iris)
And output from extract tab:
# datacleaning with datacleanr (0.0.1)
# ##------ Wed Oct 07 12:54:03 2020 ------##
library(dplyr)
library(datacleanr)
# adding column for unique IDs;
iris$.dcrkey <- seq_len(nrow(iris))
iris <- dplyr::group_by(iris, Species)
# stats and scoping level for filtering
filter_conditions <- structure(list(filter = "Sepal.Width > 2.7", grouping = list(NULL)), row.names = c(NA,
-1L), class = c("tbl_df", "tbl", "data.frame"))
# applying (scoped) filtering by groups;
iris <- datacleanr::filter_scoped_df(dframe = iris, condition_df = filter_conditions)
# observations from manual selection (Viz tab);
iris_outlier_selection <- structure(list(.dcrkey = c(15L, 16L, 19L, 34L), .annotation = c("", "", "",
"")), class = "data.frame", row.names = c(NA, -4L))
# create data set with annotation column (non-outliers are NA);
iris <- dplyr::left_join(iris, iris_outlier_selection, by = ".dcrkey")
# remove comment below to drop manually selected obs in data set;
# iris <- iris %>% dplyr::filter(is.na(.annotation))
Example 2
Launching with an .RDS from disk:
saveRDS(iris, file = "./testiris.Rds")
library(datacleanr)
dcr_app("./testiris.Rds")
Examples:
1. Exploring soil respiration with COSORE:
COSORE is a community-driven soil respiration database, recently introduced with a manuscript published here by Bond-Lamberty et al.. The database provides soil respiration flux estimates, as well as meta data across multiple data sets. Let’s explore!
remotes::install_github("bpbond/cosore")
library(dplyr)
# check data base info
db_info <- cosore::csr_database()
tibble::glimpse(db_info)
# grab one data set and explore in detail
dset <- "d20190409_ANJILELI"
anjilleli <- cosore::csr_dataset(dset)
tibble::glimpse(anjilleli$description)
datacleanr::dcr_app(anjilleli$data)
Explore sampling locations:
# Check location info
db_info <- db_info %>%
mutate(lon = CSR_LONGITUDE,
lat = CSR_LATITUDE)
datacleanr::dcr_app(db_info)
No GIF
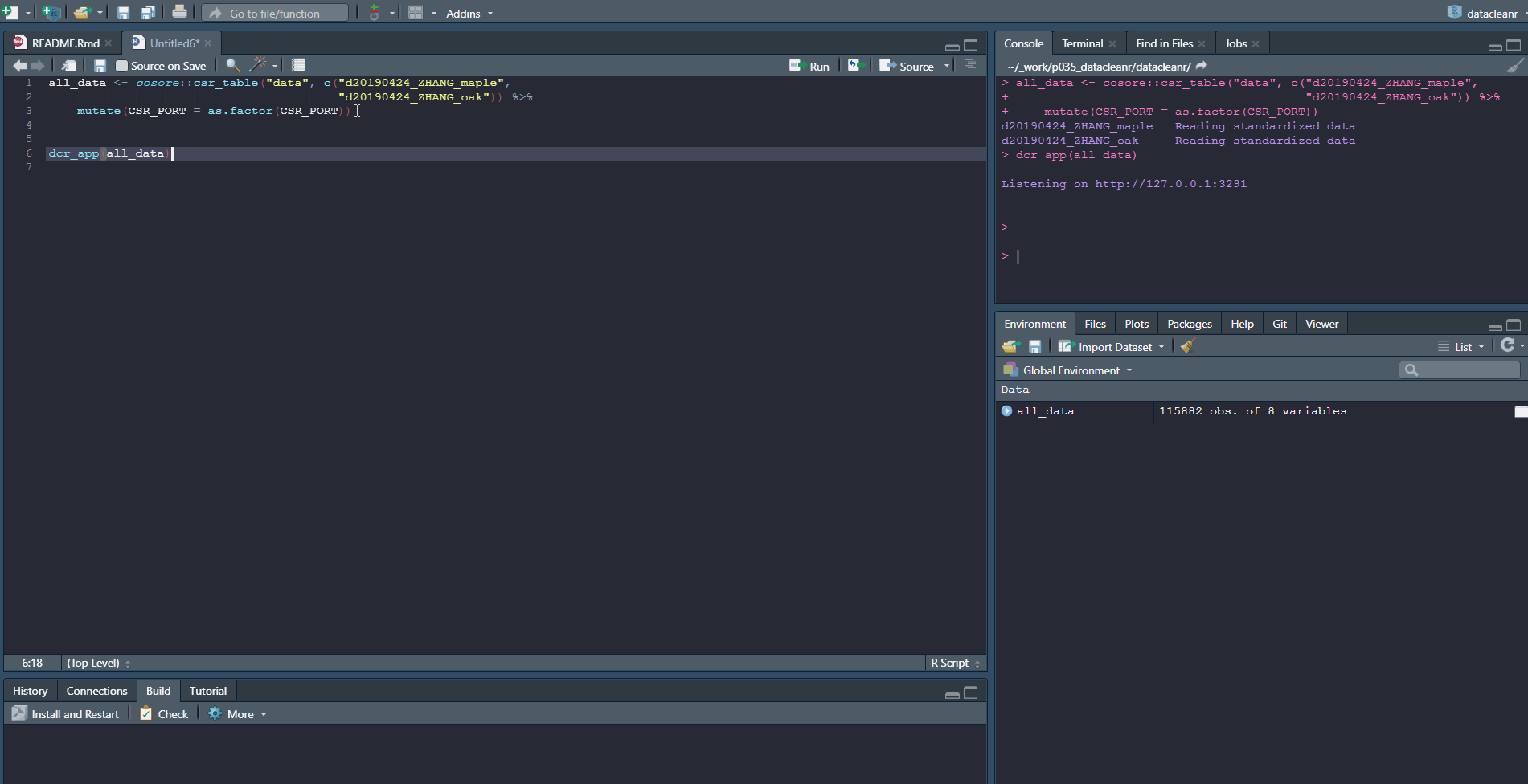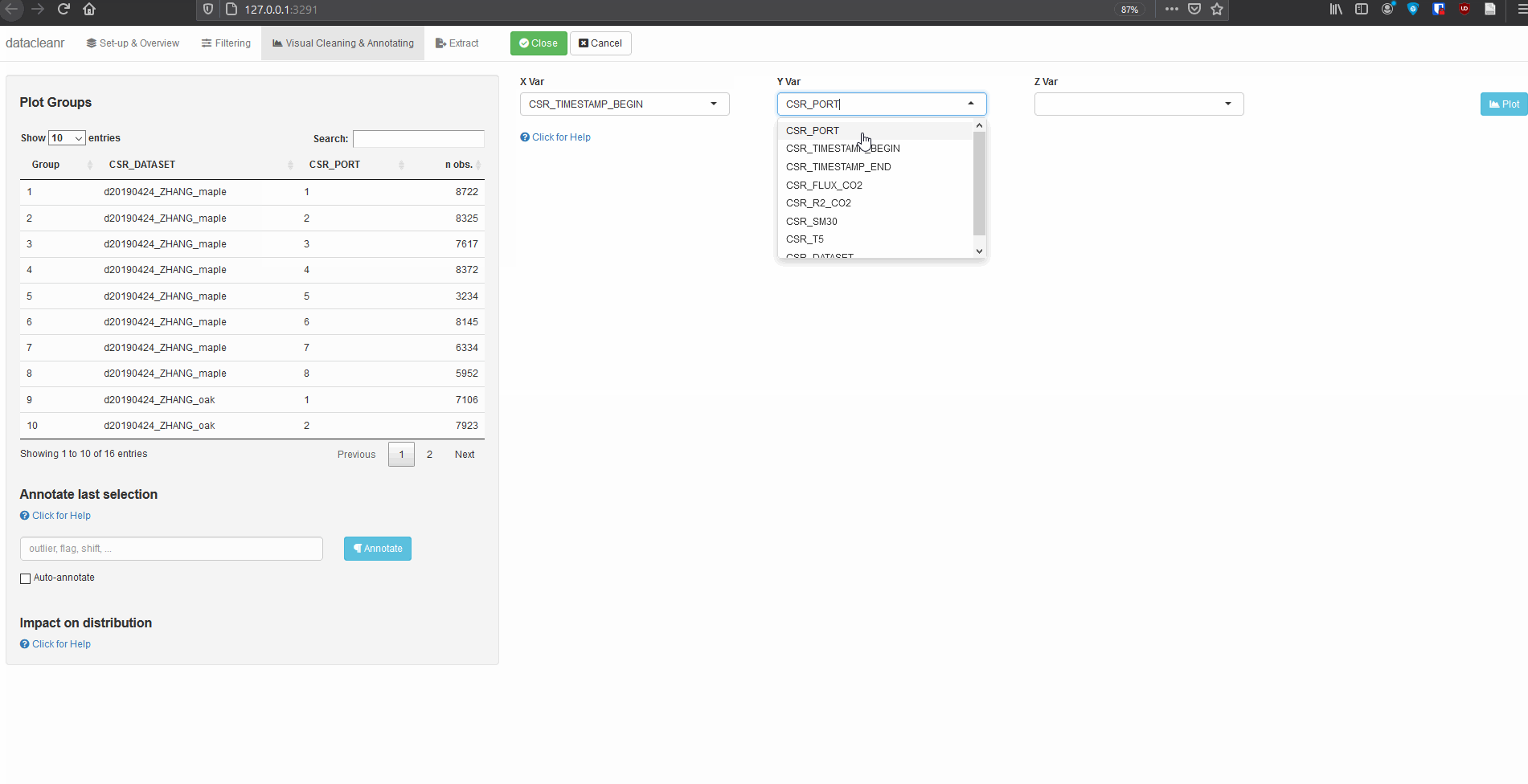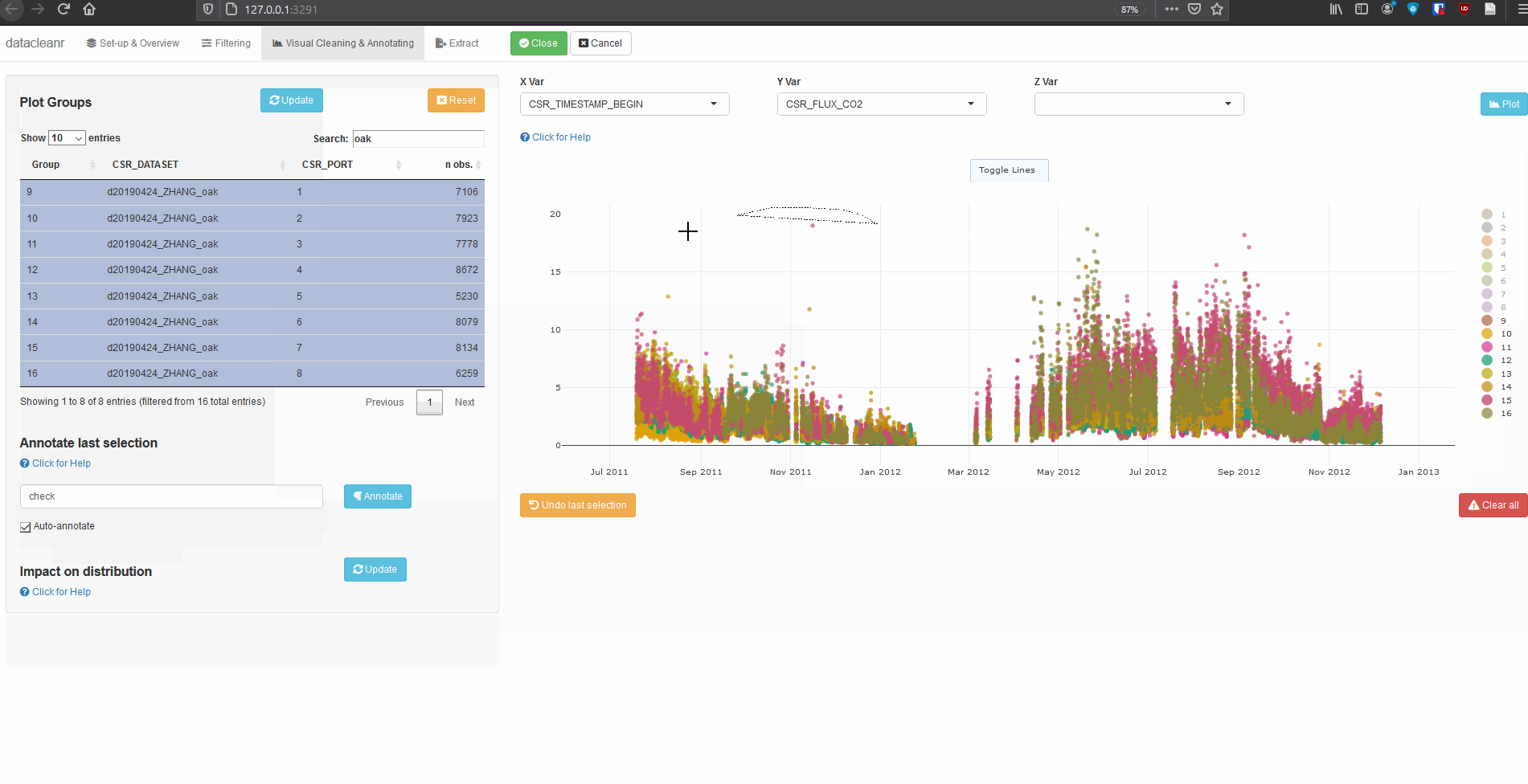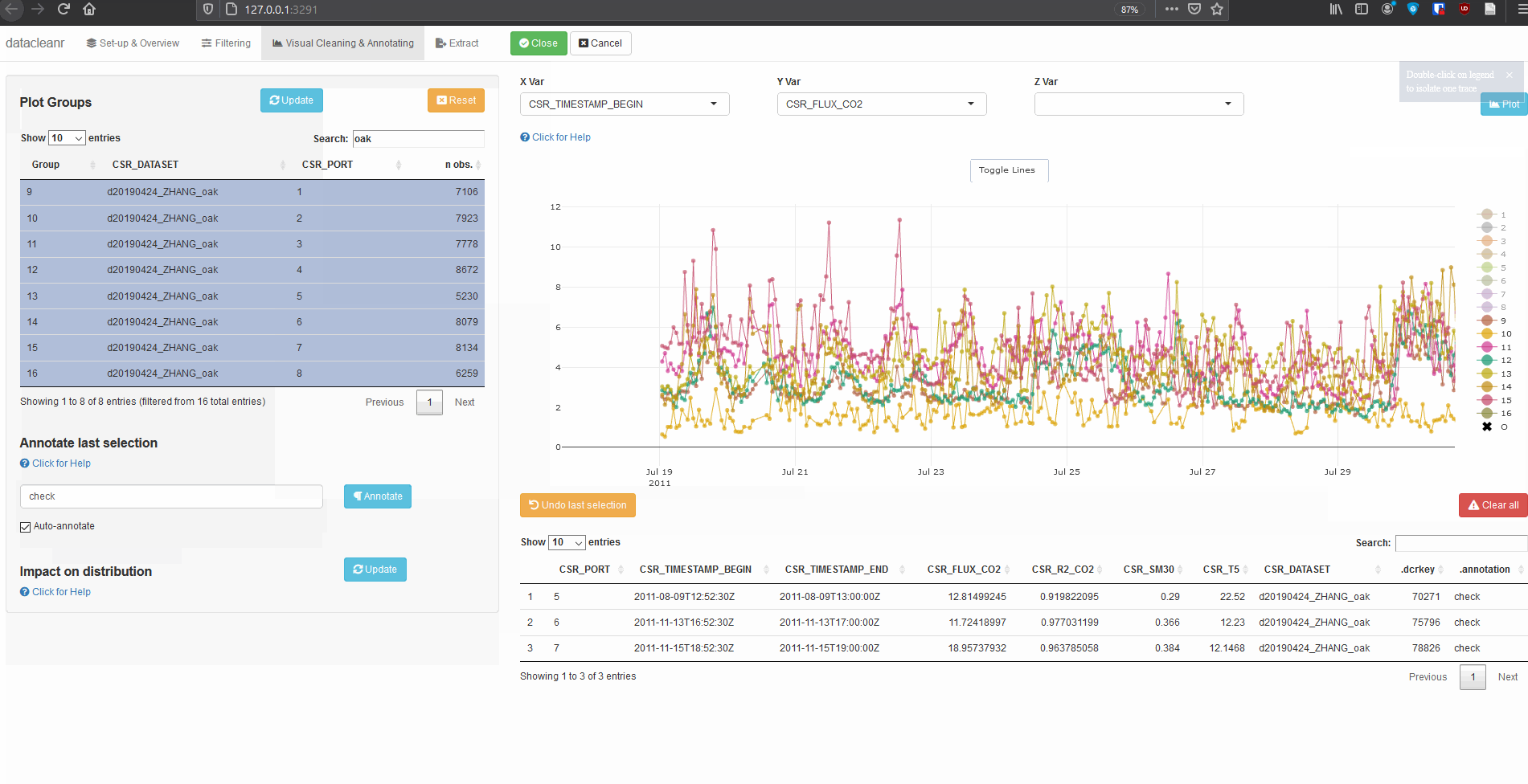
Explore nested data sets:
# grab all data from ZHANG
zhang <- cosore::csr_table("data", c("d20190424_ZHANG_maple",
"d20190424_ZHANG_oak")) %>%
# adjust for grouping
mutate(CSR_PORT = as.factor(CSR_PORT))
# group by CSR_DATASET and CSR_PORT
datacleanr::dcr_app(zhang)
Please note that the datacleanr project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
