A Lightweight, Flexible, and Fast Data Validation Package that Can Handle All Sizes of Data.
dataverifyr - A Lightweight, Flexible, and Fast Data Validation Package that Can Handle All Sizes of Data
The goal of dataverifyr is to allow a wide variety of flexible data validation checks (verifications). That means, you can specify a set of rules (R expressions) and compare any arbitrary dataset against it.
The package is built in such a way, that it adapts to your type of data and choice of data package (data.frame, data.table, tibble, arrow, or SQL connection) and chooses the right data backend automatically, this is especially handy when large or complicated datasets are involved. That way, you can concentrate on writing the rules and making sure that your data is valid rather than spending time writing boilerplate code.
The package is lightweight as all the heavy dependencies are Suggests-only, that means if you want to use data.table for the task, you don’t need to install the other packages (arrow, DBI, etc) unless you explicitly tell R to install all suggested packages as well when installing the package.
The backend for your analysis is automatically chosen based on the type of input dataset as well as the available packages. By using the underlying technologies and handing over all evaluation of code to the backend, this package can deal with all sizes of data the backends can deal with.
Installation
You can install the development version of dataverifyr like so:
# development version
# devtools::install_github("DavZim/dataverifyr")
# CRAN release
install.packages("dataverifyr")
Example
This is a basic example which shows you how to
- create a rule set manually, consisting of R expressions
- check if a dataset matches all given rules
- save and load the rules to a yaml-file for better maintainability
Note that each rule is an R expression that is evaluated within the dataset. Our first rule, for example, states that we believe all values of the mpg variable are in the range 10 to 30 (exclusive). At the moment rules work in a window/vectorized approach only, that means that a rule like this will work mpg > 10 * wt, whereas a rule like this sum(mpg) > 0 will not work as it aggregates values.
library(dataverifyr)
# define a rule set within our R code; alternatively in a yaml file
rules <- ruleset(
rule(mpg > 10 & mpg < 30), # mpg goes up to 34
rule(cyl %in% c(4, 8)), # missing 6 cyl
rule(vs %in% c(0, 1), allow_na = TRUE)
)
# print the rules
rules
#> <Verification Ruleset with 3 elements>
#> [1] 'Rule for: mpg' matching `mpg > 10 & mpg < 30` (allow_na: FALSE)
#> [2] 'Rule for: cyl' matching `cyl %in% c(4, 8)` (allow_na: FALSE)
#> [3] 'Rule for: vs' matching `vs %in% c(0, 1)` (allow_na: TRUE)
# check if the data matches our rules
res <- check_data(mtcars, rules)
res
#> name expr allow_na negate tests pass fail warn error time
#> 1: Rule for: mpg mpg > 10 & mpg < 30 FALSE FALSE 32 28 4 0.0055301189 secs
#> 2: Rule for: cyl cyl %in% c(4, 8) FALSE FALSE 32 25 7 0.0034749508 secs
#> 3: Rule for: vs vs %in% c(0, 1) TRUE FALSE 32 32 0 0.0004439354 secs
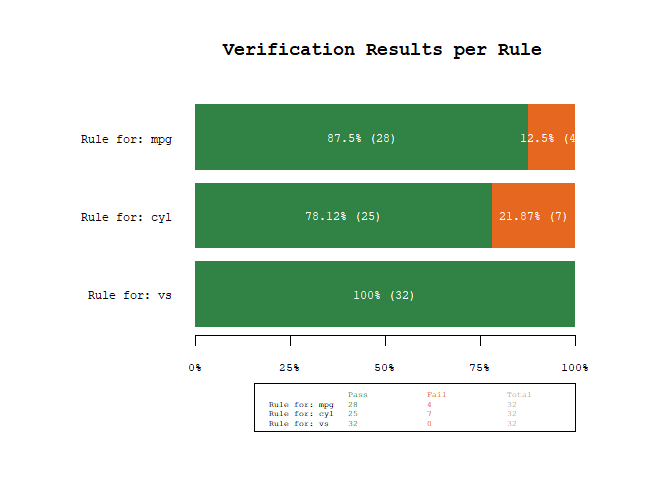
As we can see, our dataset mtcars does not conform to all of our rules. We have four fails (fail=rule is not met) for the first rule mpg > 10 & mpg < 30 (there are mpg values up to 33.9) and seven fails for the second rule cyl %in% c(4, 8) (there are cyl values of 6), while the third rule vs %in% c(0, 1) is always met.
To see which values do not meet our expectations, use the filter_fails() function
filter_fails(res, mtcars, per_rule = TRUE)
#> $`mpg > 10 & mpg < 30`
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> 1: 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
#> 2: 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> 3: 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
#> 4: 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
#>
#> $`cyl %in% c(4, 8)`
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> 1: 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
#> 2: 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
#> 3: 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
#> 4: 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
#> 5: 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
#> 6: 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
#> 7: 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
We can also visualize the results using the plot_res() function.
plot_res(res)
Note that you can also save and load a ruleset to and from a yaml file
write_rules(rules, "example_rules.yaml")
r2 <- read_rules("example_rules.yaml")
identical(rules, r2)
#> [1] TRUE
The resulting example_rules.yaml looks like this
- name: 'Rule for: mpg'
expr: mpg > 10 & mpg < 30
allow_na: no
negate: no
index: 1
- name: 'Rule for: cyl'
expr: cyl %in% c(4, 8)
allow_na: no
negate: no
index: 2
- name: 'Rule for: vs'
expr: vs %in% c(0, 1)
allow_na: yes
negate: no
index: 3
One helpful use case is to use this functionality to assert that your data has the right values in a custom read function like so:
read_custom <- function(file, rules) {
data <- read.csv(file) # or however you read in your data
# if the check_data detects a fail: the read_custom function will stop
check_data(data, rules, xname = file,
stop_on_fail = TRUE, stop_on_warn = TRUE, stop_on_error = TRUE)
# ...
data
}
# nothing happens when the data matches the rules
data <- read_custom("correct_data.csv", rules)
# an error is thrown when warnings or errors are found
data <- read_custom("wrong_data.csv", rules)
#> Error in check_data(data, rules, stop_on_fail = TRUE, stop_on_error = TRUE, stop_on_warn = TRUE) :
#> In dataset 'wrong_data.csv' found 2 rule fails, 1 warnings, 1 errors
Backends
At the moment the following backends are supported. Note that they are automatically chosen based on data type and package availability. Eg, when the dataset is a dplyr::tbl() connected to an SQLite database, the package will automatically choose RSQLite/DBI/dbplyr for the task. To see which backend dataverifyr would use for a task, you can use detect_backend(data).
| Backend / Library | Status | Data Type | Example Code | Comment |
|---|---|---|---|---|
| ✔️ |
| | When |
| ✔️ |
| | ||
| ✔️ |
| | ||
| ✔️ |
| | ||
| ✔️ |
| | Especially handy for large datasets | |
| ✔️ |
| | Note that missing values are converted to | |
| ✔️ |
| | ||
| ❓ |
| | Not tested, but should work out-of-the-box using |
Note that the rs object in the example code above refers to a ruleset(). Larger complete examples can be found below.
Larger Example using the arrow backend
For a more involved example, using a different backend, let’s say we have a larger dataset of taxi trips from NY (see also the official source of the data), that we have saved as a local arrow dataset (using parquet as a data format), where we want to make sure that some variables are in-line with our expectations/rules.
1 Download and Prepare Data
First we prepare the data by downloading it and writing the dataset to .parquet files. This needs to be done only once and is shown for reproducibility reasons only, the actual dataverifyr code is shown below the next block
library(arrow)
url <- "https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2018-01.parquet"
file <- "yellow_tripdata_2018-01.parquet"
if (!file.exists(file)) download.file(url, file, method = "curl")
file.size(file) / 1e6 # in MB
#> [1] 123.6685
# quick check of the filesize
d <- read_parquet(file)
dim(d)
#> [1] 8760687 19
names(d)
#> [1] "VendorID" "tpep_pickup_datetime" "tpep_dropoff_datetime" "passenger_count"
#> [5] "trip_distance" "RatecodeID" "store_and_fwd_flag" "PULocationID"
#> [9] "DOLocationID" "payment_type" "fare_amount" "extra"
#> [13] "mta_tax" "tip_amount" "tolls_amount" "improvement_surcharge"
#> [17] "total_amount" "congestion_surcharge" "airport_fee"
# write the dataset to disk
write_dataset(d, "nyc-taxi-data")
2 Create Rules in yaml
Next, we can create some rules that we will use to check our data. As we saw earlier, we can create the rules in R using the rule() and ruleset() functions, there is however, the (in my opinion) preferred option to separate the code from the rules by writing the rules in a separate yaml file and reading them into R.
First we display the hand-written contents of the nyc_data_rules.yaml file.
- name: 'Rule for: passenger_count'
expr: passenger_count >= 0 & passenger_count <= 10
allow_na: no
negate: no
index: 1
- name: 'Rule for: trip_distance'
expr: trip_distance >= 0 & trip_distance <= 1000
allow_na: no
negate: no
index: 2
- name: 'Rule for: payment_type'
expr: payment_type %in% c(0, 1, 2, 3, 4)
allow_na: no
negate: no
index: 3
Then, we can load, display, and finally check the rules against the data
rules <- read_rules("nyc_data_rules.yaml")
rules
#> <Verification Ruleset with 3 elements>
#> [1] 'Rule for: passenger_count' matching `passenger_count >= 0 & passenger_count <= 10` (allow_na: FALSE)
#> [2] 'Rule for: trip_distance' matching `trip_distance >= 0 & trip_distance <= 1000` (allow_na: FALSE)
#> [3] 'Rule for: payment_type' matching `payment_type %in% c(0, 1, 2, 3, 4)` (allow_na: FALSE)
3 Verify that the Data matches the given Rules
Now we can check if the data follows our rules or if we have unexpected data points:
# open the dataset
ds <- open_dataset("nyc-taxi-data/")
# perform the data validation check
res <- check_data(ds, rules)
res
#> # A tibble: 3 × 10
#> name expr allow…¹ negate tests pass fail warn error time
#> <chr> <chr> <lgl> <lgl> <int> <int> <int> <chr> <chr> <drt>
#> 1 Rule for: passenger_count passenger_count … FALSE FALSE 8760687 8760687 0 "" "" 0.56…
#> 2 Rule for: trip_distance trip_distance >=… FALSE FALSE 8760687 8760686 1 "" "" 0.43…
#> 3 Rule for: payment_type payment_type %in… FALSE FALSE 8760687 8760687 0 "" "" 0.42…
#> # … with abbreviated variable name ¹allow_na
plot_res(res)
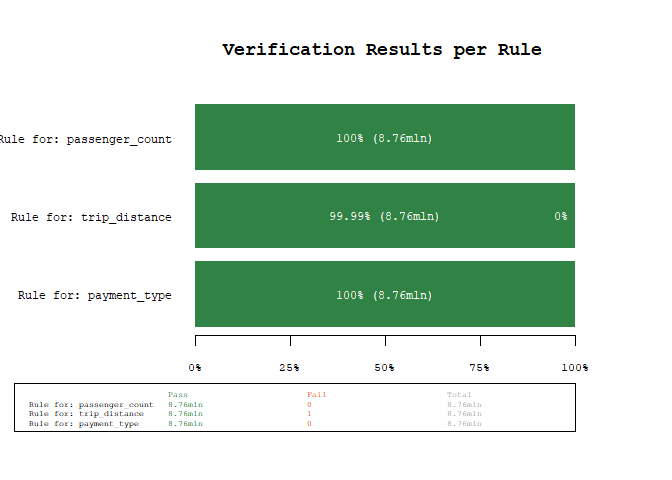
Using the power of arrow, we were able to scan 8+mln observations for three rules in about 1.5 seconds (YMMV). As we can see from the results, there is one unexpected value, lets quickly investigate using the filter_fails() function, which filters a dataset for the failed rule matches
res |>
filter_fails(ds) |>
# only select a couple of variables for brevity
dplyr::select(tpep_pickup_datetime, tpep_dropoff_datetime, trip_distance)
#> # A tibble: 1 × 3
#> tpep_pickup_datetime tpep_dropoff_datetime trip_distance
#> <dttm> <dttm> <dbl>
#> 1 2018-01-30 12:41:02 2018-01-30 12:42:09 189484.
As we can see, this is probably a data error (a trip distance of 190k miles in 1 minute seems - ehm stellar…).
Using a DBI Backend
If you have a SQLite or duckdb database, you can use the package like this
library(DBI)
library(dplyr)
# connect to a duckdb database
con <- dbConnect(duckdb::duckdb("duckdb-database.duckdb"))
# for demo purposes write the data once
dbWriteTable(con, "mtcars", mtcars)
# create a tbl connection, which can be used in the checks
tbl <- tbl(con, "mtcars")
# create rules
rules <- ruleset(
rule(mpg > 10 & mpg < 30),
rule(cyl %in% c(4, 8)),
rule(vs %in% c(0, 1), allow_na = TRUE)
)
# check rules
res <- check_data(tbl, rules)
res
#> # A tibble: 3 × 10
#> name expr allow_na negate tests pass fail warn error time
#> <chr> <chr> <lgl> <lgl> <dbl> <dbl> <dbl> <chr> <chr> <drtn>
#> 1 Rule for: mpg mpg > 10 & mpg < 30 FALSE FALSE 32 28 4 "" "" 1.232728 secs
#> 2 Rule for: cyl cyl %in% c(4, 8) FALSE FALSE 32 25 7 "" "" 0.2015200 secs
#> 3 Rule for: vs vs %in% c(0, 1) TRUE FALSE 32 32 0 "" "" 0.1898661 secs
filter_fails(res, tbl, per_rule = TRUE)
#> $`mpg > 10 & mpg < 30`
#> # A tibble: 4 × 11
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 32.4 4 78.7 66 4.08 2.2 19.47 1 1 4 1
#> 2 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> 3 33.9 4 71.1 65 4.22 1.835 19.9 1 1 4 1
#> 4 30.4 4 95.1 113 3.77 1.513 16.9 1 1 5 2
#>
#> $`cyl %in% c(4, 8)`
#> # A tibble: 7 × 11
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 21 6 160 110 3.9 2.62 16.46 0 1 4 4
#> 2 21 6 160 110 3.9 2.875 17.02 0 1 4 4
#> 3 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
#> 4 18.1 6 225 105 2.76 3.46 20.22 1 0 3 1
#> 5 19.2 6 167.6 123 3.92 3.44 18.3 1 0 4 4
#> 6 17.8 6 167.6 123 3.92 3.44 18.9 1 0 4 4
#> 7 19.7 6 145 175 3.62 2.77 15.5 0 1 5 6
# lastly disconnect from the database again
dbDisconnect(con, shutdown = TRUE)
Alternative Data Validation R Libraries
If this library is not what you are looking for, the following might be good alternatives to validate your data: