Polygons of Bivariate Density Distributions.
densityarea 
The package {ggdensity}[^1] allows for plotting interpretable bivariate densities by using highest density ranges (HDRs). For example:
library(tibble)
library(ggplot2)
library(ggdensity)
set.seed(10)
df <- tibble(
x = c(rnorm(100), rnorm(100, mean = 3)),
y = c(rnorm(100), rnorm(100, mean = 3))
)
ggplot(df, aes(x,y))+
stat_hdr()
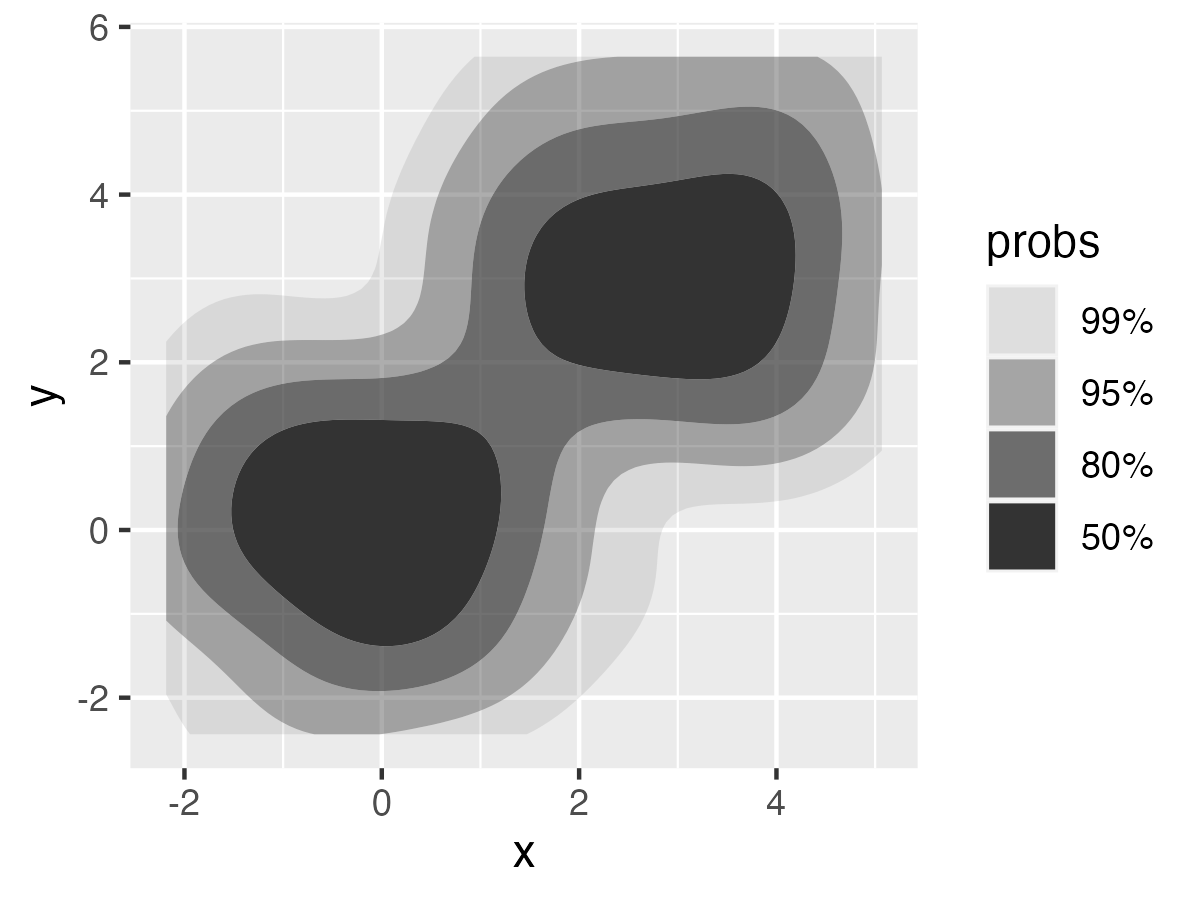
{densityarea} gives direct access to these HDRs, either as data frames or as simple features, for further analysis.
Installation
You can install {densityarea} from CRAN with:
install.packages("densityarea")
Or you can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("JoFrhwld/densityarea")
Example
The use case the package was initially developed for was for estimating vowel space areas.
library(densityarea)
library(dplyr)
library(tidyr)
library(sf)
data(s01)
# initial data processing
s01 |>
mutate(lF1 = -log(F1),
lF2 = -log(F2))->
s01
To get this speaker’s vowel space area we can pass the data through dplyr::reframe()
s01 |>
reframe(
density_area(lF2, lF1, probs = 0.8)
)
#> # A tibble: 1 × 3
#> level_id prob area
#> <int> <dbl> <dbl>
#> 1 1 0.8 0.406
Or, we could get the spatial polygon associated with the 80% probability level
s01 |>
reframe(
density_polygons(lF2, lF1, probs = 0.8, as_sf = T)
)
#> # A tibble: 1 × 3
#> level_id prob geometry
#> <int> <dbl> <POLYGON>
#> 1 1 0.8 ((-7.777586 -6.009484, -7.801131 -6.010429, -7.824676 -6.01700…
For more
For more details on using {densityarea}, see , and for further information on using spatial polygons, see vignette("sf-operations").
[^1]: Otto J, Kahle D (2023). ggdensity: Interpretable Bivariate Density Visualization with ‘ggplot2’. <https://github.com/jamesotto852/ggdensity/