Power Analysis for Differential Expression Studies.
depower
Overview
depower provides a convenient framework to simulate, test, power, and visualize data for differential expression studies with lognormal or negative binomial outcomes. Supported designs are two-sample comparisons of both independent and dependent outcomes. Power may be summarized in the context of controlling the per-family error rate or family-wise error rate.
Installation
# Install from CRAN
install.packages("depower")
# Or the development version from bitbucket
remotes::install_bitbucket("bklamer/depower")
Usage
library(depower)
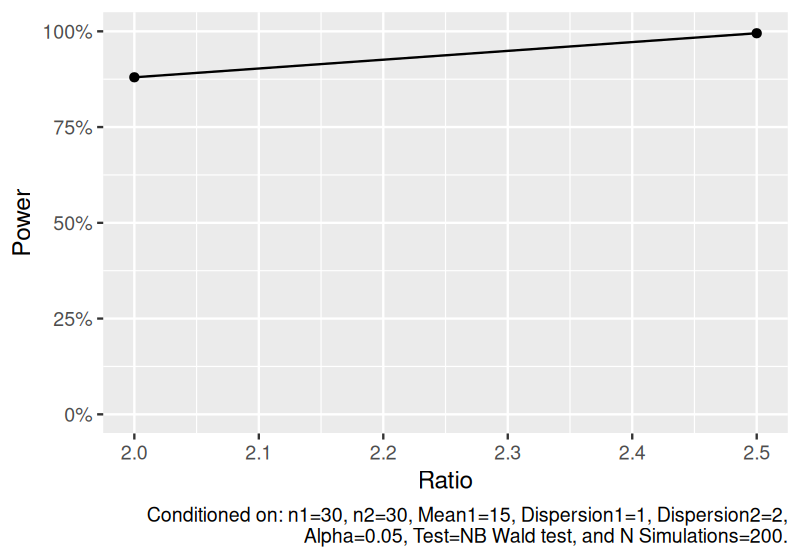
Estimate power to detect the ratio of means for independent two-sample negative binomial data.
set.seed(1234)
power_nb <- sim_nb(
n1 = 30,
n2 = 30,
mean1 = 15,
ratio = c(2, 2.5),
dispersion1 = 1,
dispersion2 = 2,
nsims = 200
) |>
power()
power_nb
#> # A tibble: 2 × 12
#> n1 n2 mean1 mean2 ratio dispersion1 dispersion2 distribution nsims test alpha
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
#> 1 30 30 15 30 2 1 2 Independent … 200 NB W… 0.05
#> 2 30 30 15 37.5 2.5 1 2 Independent … 200 NB W… 0.05
#> # ℹ 1 more variable: power <dbl>
plot(power_nb)
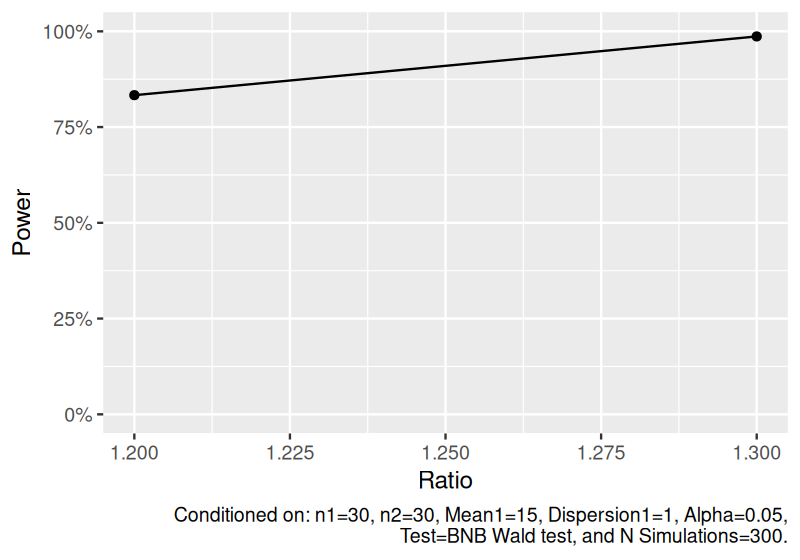
Estimate power to detect the ratio of means for bivariate negative binomial data.
set.seed(1234)
power_bnb <- sim_bnb(
n = 30,
mean1 = 15,
ratio = c(1.2, 1.3),
dispersion = 1,
nsims = 300
) |>
power()
power_bnb
#> # A tibble: 2 × 11
#> n1 n2 mean1 mean2 ratio dispersion1 distribution nsims test alpha power
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl>
#> 1 30 30 15 18 1.2 1 Dependent two-samp… 300 BNB … 0.05 0.833
#> 2 30 30 15 19.5 1.3 1 Dependent two-samp… 300 BNB … 0.05 0.987
plot(power_bnb)
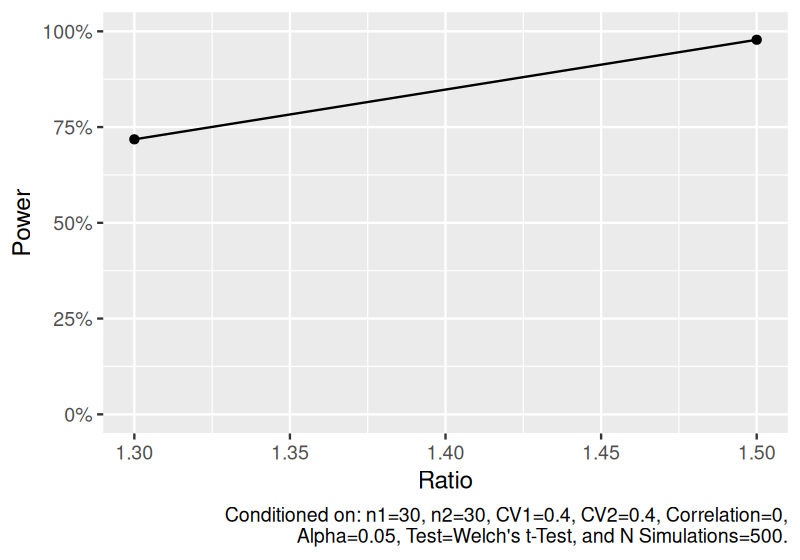
Estimate power to detect the geometric ratio of means for independent two-sample lognormal data.
set.seed(1234)
power_ind_lognormal <- sim_log_lognormal(
n1 = 30,
n2 = 30,
ratio = c(1.3, 1.5),
cv1 = 0.4,
cv2 = 0.4,
nsims = 500
) |>
power()
power_ind_lognormal
#> # A tibble: 2 × 11
#> n1 n2 ratio cv1 cv2 cor distribution nsims test alpha power
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl>
#> 1 30 30 1.3 0.4 0.4 0 Independent two-sample l… 500 Welc… 0.05 0.718
#> 2 30 30 1.5 0.4 0.4 0 Independent two-sample l… 500 Welc… 0.05 0.978
plot(power_ind_lognormal)
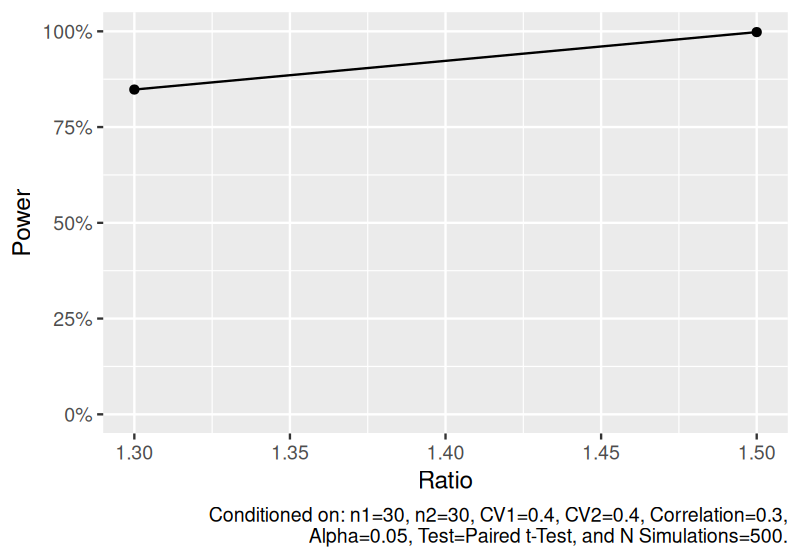
Estimate power to detect the geometric mean ratio for dependent two-sample lognormal data.
set.seed(1234)
power_dep_lognormal <- sim_log_lognormal(
n1 = 30,
n2 = 30,
ratio = c(1.3, 1.5),
cv1 = 0.4,
cv2 = 0.4,
cor = 0.3,
nsims = 500
) |>
power()
power_dep_lognormal
#> # A tibble: 2 × 11
#> n1 n2 ratio cv1 cv2 cor distribution nsims test alpha power
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl>
#> 1 30 30 1.3 0.4 0.4 0.3 Dependent two-sample log… 500 Pair… 0.05 0.848
#> 2 30 30 1.5 0.4 0.4 0.3 Dependent two-sample log… 500 Pair… 0.05 0.998
plot(power_dep_lognormal)
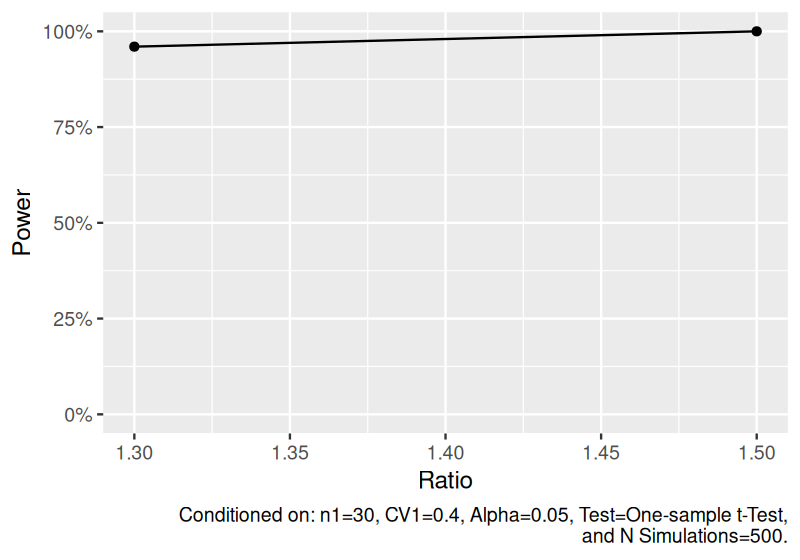
Estimate power to detect the geometric mean for one-sample lognormal data.
set.seed(1234)
power_one_lognormal <- sim_log_lognormal(
n1 = 30,
ratio = c(1.3, 1.5),
cv1 = 0.4,
nsims = 500
) |>
power()
power_one_lognormal
#> # A tibble: 2 × 8
#> n1 ratio cv1 distribution nsims test alpha power
#> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl>
#> 1 30 1.3 0.4 One-sample log(lognormal) 500 One-sample t-Test 0.05 0.96
#> 2 30 1.5 0.4 One-sample log(lognormal) 500 One-sample t-Test 0.05 1
plot(power_one_lognormal)