Diewert and Fox's Method of Value Added Growth Decomposition.
dfvad
Overview
dfvad decomposes value added growth into explanatory factors. A cost constrained value added function is defined to specify the production frontier. Industry estimates can also be aggregated using a weighted average approach.
Installation
dfvad is available from https://github.com/shipei-zeng/dfvad. To install it, install_github from the devtools package can be helpful.
devtools::install_github("shipei-zeng/dfvad")
If error messages show that the URL cannot be opened, please set the download option before installing it.
options(download.file.method = "libcurl")
If error messages show that schannel failed to receive handshake, please delete the previous package before updating it.
It has also been uploaded to the CRAN repository, which can be downloaded using:
install.packages("dfvad")
Usage
value_decom() for decomposing nominal value added growth identifies the contributions from efficiency change, growth of primary inputs, changes in output and input prices, technical progress and returns to scale.
library(dfvad)
# Use the built-in dataset "mining"
table1 <- value_decom(c("h2","x2"), c("w2","u2"), "y2", "p2", "year", mining)[[1]]
head(table1)
#> period value alpha beta gamma efficiency epsilon tau
#> 1 1991 1.0869517 1.0287049 0.9944262 1.0000000 1.0000000 1.0000000 1.062544
#> 2 1992 0.9960608 0.9523963 0.9874494 1.0000000 1.0000000 1.0000000 1.059140
#> 3 1993 1.0477108 1.0373754 1.0180111 0.9997303 0.9923619 0.9923619 1.000000
#> 4 1994 0.9773035 0.9605188 1.0444275 0.9996838 0.9670585 0.9745018 1.000000
#> 5 1995 1.0545680 0.9842271 1.0168128 0.9999874 1.0000000 1.0340636 1.019052
#> 6 1996 1.1345729 1.0406754 1.0327427 1.0000000 1.0000000 1.0000000 1.055662
#> TFPG
#> 1 1.0625440
#> 2 1.0591398
#> 3 0.9920943
#> 4 0.9741937
#> 5 1.0537516
#> 6 1.0556623
table2 <- value_decom(c("h2","x2"), c("w2","u2"), "y2", "p2", "year", mining)[[2]]
head(table2)
#> period value A B C E T TFP
#> 1 1990 1.000000 1.0000000 1.0000000 1.0000000 1.0000000 1.000000 1.000000
#> 2 1991 1.086952 1.0287049 0.9944262 1.0000000 1.0000000 1.062544 1.062544
#> 3 1992 1.082670 0.9797347 0.9819455 1.0000000 1.0000000 1.125383 1.125383
#> 4 1993 1.134325 1.0163527 0.9996314 0.9997303 0.9923619 1.125383 1.116486
#> 5 1994 1.108580 0.9762259 1.0440425 0.9994142 0.9670585 1.125383 1.087673
#> 6 1995 1.169073 0.9608280 1.0615958 0.9994016 1.0000000 1.146824 1.146138
t_weight() follows a “bottom up” approach that uses weighted averages of the sectoral decompositions to provide an approximate decomposition into explanatory components at the aggregate level.
library(dfvad)
# Use the built-in dataset "sector"
table1 <- t_weight("y", "p", "industry", "year", "alpha", "beta", "gamma", "epsilon", "tau", sector)[[1]]
head(table1)
#> period value alpha beta gamma epsilon tau TFPG
#> 1 1991 0.9951654 1.004989 0.9878890 0.9996087 0.9868727 1.016024 1.0023647
#> 2 1992 1.0145281 1.015884 0.9869371 0.9987753 0.9962747 1.016905 1.0118834
#> 3 1993 1.0656435 1.034633 1.0128698 1.0002431 1.0011899 1.015434 1.0168858
#> 4 1994 1.0649234 1.007479 1.0291213 1.0001043 1.0029142 1.024013 1.0271072
#> 5 1995 1.0565961 1.020031 1.0378505 1.0010086 0.9871089 1.010072 0.9980697
#> 6 1996 1.0703334 1.019766 1.0212297 0.9998535 1.0097033 1.018044 1.0277682
table2 <- t_weight("y", "p", "industry", "year", "alpha", "beta", "gamma", "epsilon", "tau", sector)[[2]]
head(table2)
#> period value A B C E T TFP
#> 1 1990 1.0000000 1.000000 1.0000000 1.0000000 1.0000000 1.000000 1.000000
#> 2 1991 0.9951654 1.004989 0.9878890 0.9996087 0.9868727 1.016024 1.002365
#> 3 1992 1.0096232 1.020952 0.9749843 0.9983845 0.9831963 1.033200 1.014276
#> 4 1993 1.0758984 1.056311 0.9875322 0.9986272 0.9843662 1.049147 1.031403
#> 5 1994 1.1457494 1.064211 1.0162904 0.9987313 0.9872349 1.074340 1.059361
#> 6 1995 1.2105943 1.085528 1.0547575 0.9997387 0.9745083 1.085161 1.057317
Display
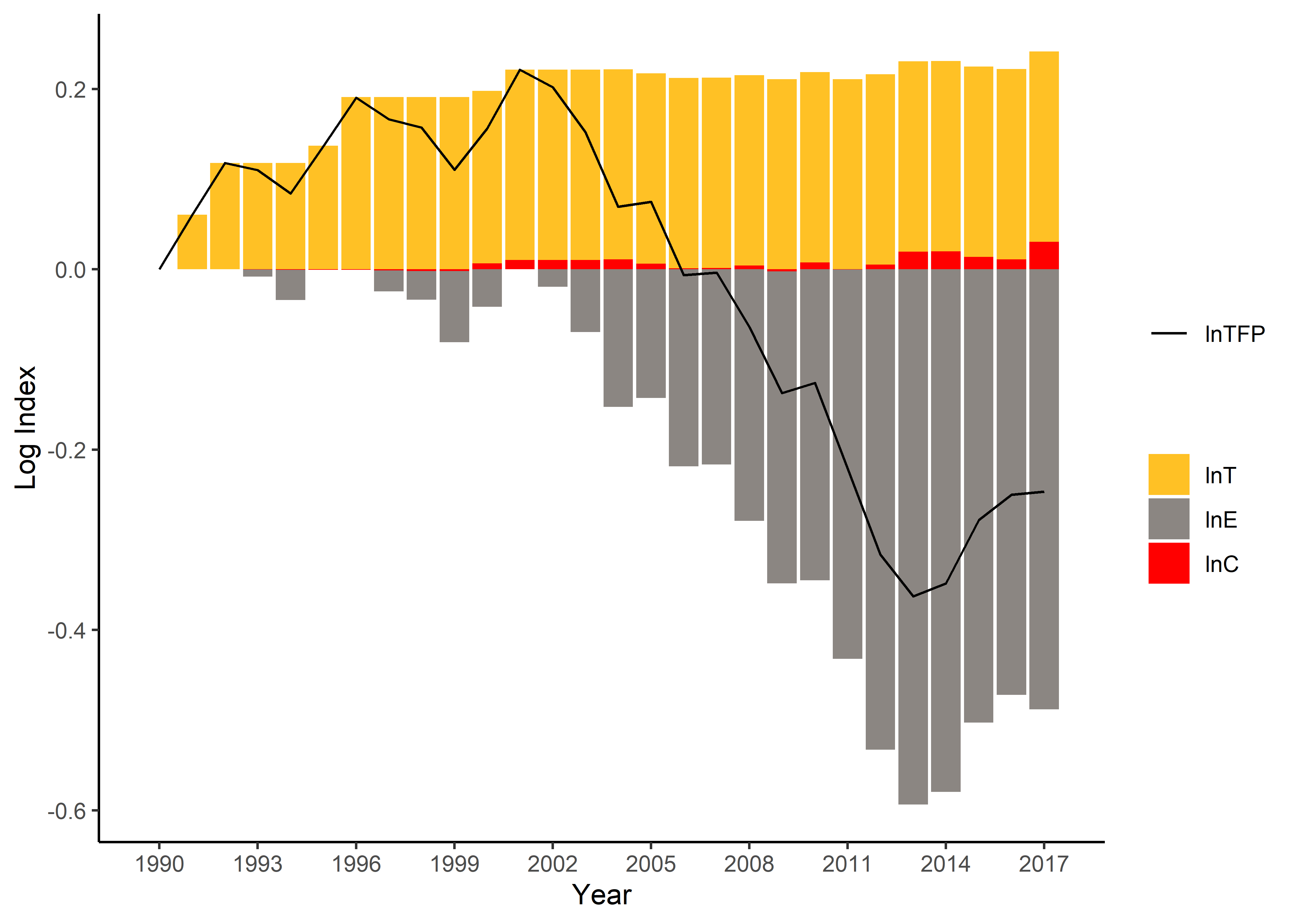
Here is an example to plot the explanatory factors of productivity (logarithmic indexes). Additional packages such as ggplot2 and reshape2 are required.
library(dfvad)
library(ggplot2)
library(reshape2)
# Get the decomposition result
df <- value_decom(c("h2","x2"), c("w2","u2"), "y2", "p2", "year", mining)[[2]]
# Extract columns and rename
df_cmpt <- data.frame(df[,"period"], log(df[,c("T", "E", "C")]))
colnames(df_cmpt) <- c("year", "lnT", "lnE", "lnC")
df_tfp <- data.frame(df[,"period"], log(df[,"TFP"]))
colnames(df_tfp) <- c("year", "lnTFP")
# Set the colour scheme
palette_a <- c("goldenrod1", "seashell4", "red")
# Convert data into a tidy form
df_cmpt_tidy <- melt(df_cmpt, id.vars="year")
# Plot the components
plot_out <- ggplot(df_cmpt_tidy) + geom_bar(aes(x=year, y=value, fill=variable), stat="identity") +
geom_line(data=df_tfp, aes(x=year,y=lnTFP,color='black'), lwd=0.5) +
ylab('Log Index') + xlab('Year') +
scale_fill_manual("", values=palette_a) +
scale_colour_manual("", values=c('black'='black'), labels = c('lnTFP')) +
scale_x_continuous(breaks = seq(min(df$period), max(df$period), by = 3)) +
theme_classic()
print(plot_out)