Species Distribution and Abundance Modelling at High Spatio-Temporal Resolution.
dynamicSDM
Added features dynamicSDM v1.3
Added function
extract_static_coordsfor extracting spatially-buffered co-ordinate data from static datasets.Added arguments to
dynamic_proj_covariates()for adding static rasters to covariates for each data (e.g. static elevation raster)Removed package dependency on
raster,sp,geodistandgeosphere.All functions are now
terraandsfcompatible.The package has since been published in the Open Access journal “Methods in Ecology and Evolution”
Dobson, R., Challinor, A.J., Cheke, R.A., Jennings, S., Willis, S.G. and Dallimer, M., 2023. dynamicSDM: An R package for species geographical distribution and abundance modelling at high spatiotemporal resolution. Methods in Ecology and Evolution, 14, 1190-1199.
Summary
Across ecological research fields, species distribution and abundance modelling (SDM) is a major tool for understanding the drivers and patterns of species occurrence. To advance our ability to model species inhabiting dynamic ecosystems worldwide, dynamicSDM facilitates the incorporation of explanatory variables that are dynamic in both space and time. Our functions are:
- user-friendly - requiring only simple inputs and outputs;
- highly flexible - offering diverse and open arguments for targeting study specifics;
- computer friendly - utilising Google Earth Engine and Google Drive to minimise the computing power and storage demands associated with high spatiotemporal resolution modelling.
Package structure
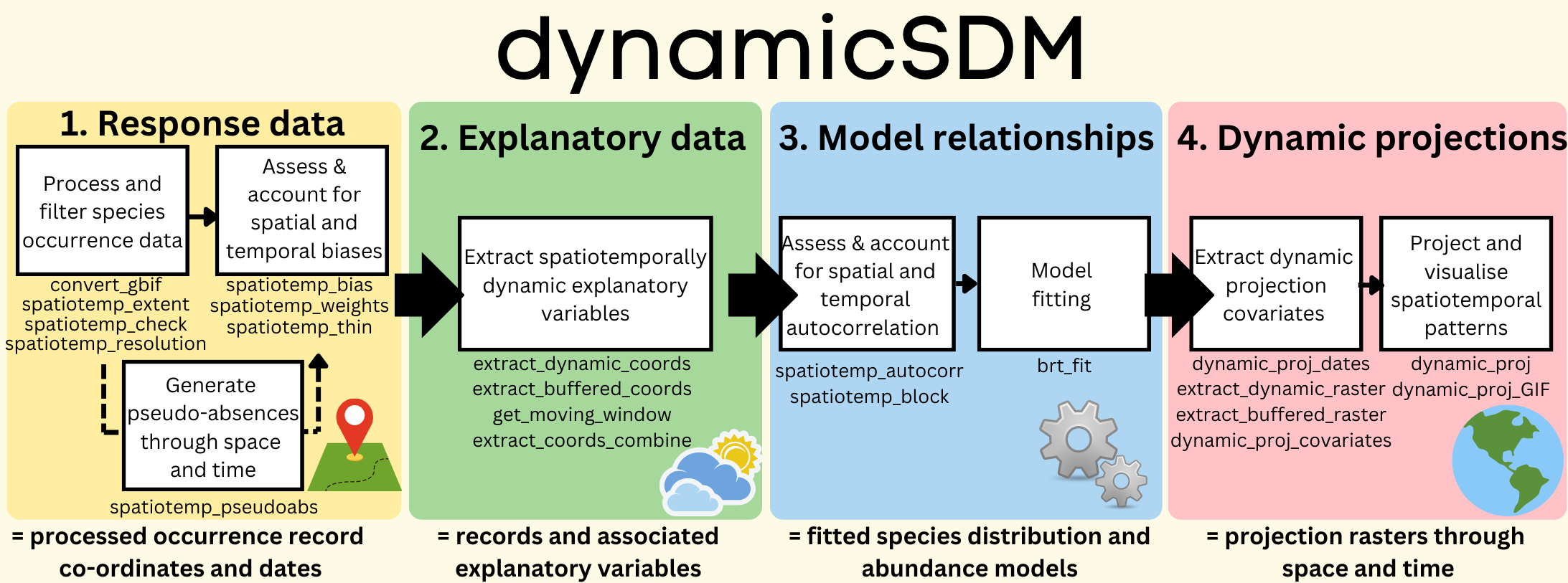
dynamicSDM functions are split into four key modelling stages: response data, explanatory variables, modelling relationships and dynamic projections. See the package manual here for more details on each function.
1) Response data functions
Functions for preparing species distribution or abundance model input data for modelling with spatiotemporally dynamic explanatory variables.
convert_gbif()Transform Global Biodiversity Information Facility occurrence records todynamicSDMcompatible.spatiotemp_check()Check species occurrence record formatting, completeness and validity.spatiotemp_extent()Filter species occurrence records by a given spatial and temporal extent.spatiotemp_resolution()Filter species occurrence records by given spatial and temporal resolution.spatiotemp_bias()Test for spatial and temporal bias in species occurrence records.spatiotemp_thin()Thin species occurrence records by spatial and temporal proximity.spatiotemp_pseudoabs()Generate pseudo-absence record coordinates and dates.spatiotemp_weights()Calculate sampling effort across spatial and temporal buffer from occurrence records.
2) Explanatory variable functions
Functions for extracting spatiotemporally dynamic explanatory variable data for species occurrence record co-ordinates and dates using Google Earth Engine.
extract_dynamic_coords()Extract temporally dynamic explanatory variable data for occurrence records.get_moving_window()Generate a “moving window” matrix of optimal size for spatial buffering of explanatory variable data.extract_buffered_coords()Extract spatially buffered and temporally dynamic explanatory variable data for occurrence records.extract_coords_combine()Combine extracted explanatory variable data for occurrence records into single data frame for model fitting.extract_static_coordsExtract spatially buffered data from static rasters for occurrence record co-ordinates (no temporal dimension).
3) Modelling relationship functions
Functions for generating species distribution or abundance models that account for spatial and temporal autocorrelation in dynamic explanatory variables.
spatiotemp_autocorr()Test for spatial and temporal autocorrelation in species distribution model explanatory data.spatiotemp_block()Split occurrence records into spatial and temporal blocks for model fitting.brt_fit()Fit boosted regression tree models to species distribution or abundance data.
4) Dynamic projection functions
Functions for generating explanatory variable projection data frames at given spatiotemporal extent and resolution, and projecting species dynamic distribution and abundance patterns onto these.
dynamic_proj_dates()Generate vector of dates for dynamic projectionsextract_dynamic_raster()Extract temporally dynamic rasters of explanatory variable data.extract_buffered_raster()Extract spatially buffered and temporally dynamic rasters of explanatory variable data.dynamic_proj_covariates()Combine explanatory variable rasters into a covariate data frame for each projection date.dynamic_proj()Project species distribution and abundance models onto dynamic environmental covariates.dynamic_proj_GIF()Create GIF of dynamic species distribution and abundance projections
Installation
# Install using Github
install_github("r-a-dobson/dynamicSDM")
Common installation errors
dynamicSDM depends on a range of spatial and graphic R packages, which may result in some persistent errors on installation or running of certain functions.
If you encounter an error or bug when installing and using dynamicSDM, please post a comment here for guidance and support from us.
Below we have outlined common errors and typical solutions to try, depending on your operating system
1) Error with rgl
# Loading rgl's DLL failed. This build of rgl depends on XQuartz, which failed to load.
options(rgl.useNULL = TRUE)
library(rgl)
2) Dependency package terra
On Homebrew (macOS) run:
brew install pkg-config
brew install gdal
On Linux run:
sudo apt-get install libgdal-dev libproj-dev libgeos-dev libudunits2-dev netcdf-bin
Then in R run:
install.packages("Rcpp")
install.packages('terra', repos='https://rspatial.r-universe.dev')
3) Dependency package magick
On Homebrew (macOS) run:
brew install imagemagick@6
On Linux run:
sudo apt-get install -y libmagick++-dev
Then in R run:
install.packages("magick")