A Multi-Modal Simulator for Spearheading Single-Cell Omics Analyses.
dyngen 
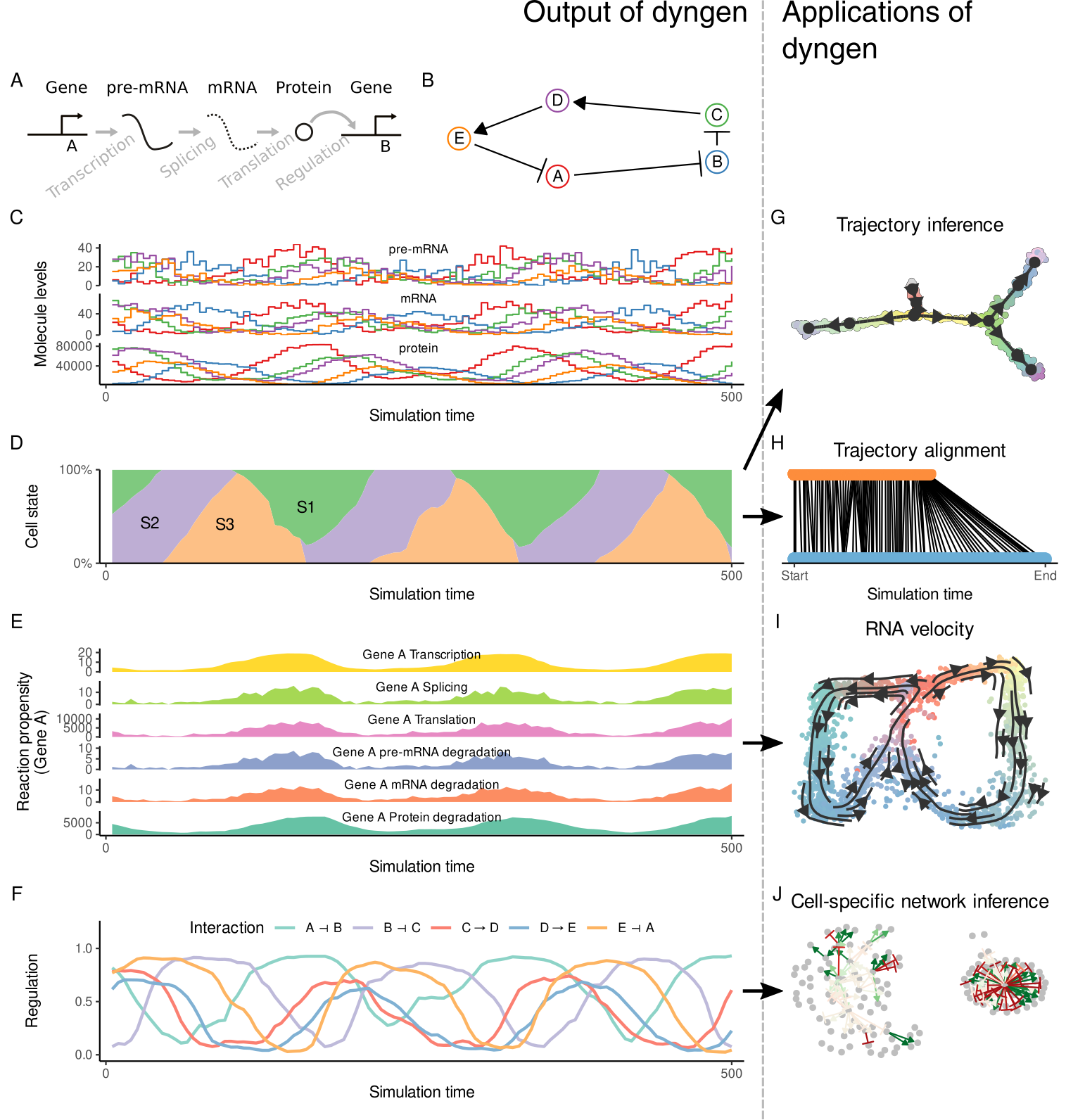
dyngen is a novel, multi-modal simulation engine for studying dynamic cellular processes at single-cell resolution. dyngen is more flexible than current single-cell simulation engines, and allows better method development and benchmarking, thereby stimulating development and testing of novel computational methods.
dyngen is now published (CC-BY, doi:10.1038/s41467-021-24152-2). Run citation("dyngen") to obtain the corresponding citation information. All source code for reproducing the results in this manuscript are available on GitHub.
Installation
dyngen should work straight out of the CRAN box by running install.packages("dyngen"). Having said that, you should definitely configure a few system variables for optimal speed. Check the installation guide for more information!
Getting started
Check out this guide on how to get started with dyngen. You can find more guides by clicking any of the links below:
- Getting started
- Installation instructions
- Showcase different backbones
- Advanced: Comparison to reference dataset
- Advanced: Constructing a custom backbone
- Advanced: Running dyngen from a docker container
- Advanced: On scalability and runtime
- Advanced: Simulating batch effects
- Advanced: Simulating a knockout experiment
- Advanced: Tweaking parameters
Latest changes
A full list of changes is available on our changelog.
