Provides an R Interface to 'Enrichr'.
An R interface to the Enrichr database
Wajid Jawaid 2025-02-02
Installation
enrichR can be installed from Github or from CRAN.
Github
library(devtools)
install_github("wjawaid/enrichR")
CRAN
The package can be downloaded from CRAN using:
install.packages("enrichR")
Usage example
enrichR provides an interface to the Enrichr database (Kuleshov et al. 2016) hosted at https://maayanlab.cloud/Enrichr/.
By default human genes are selected otherwise select your organism of choice. (This functionality was contributed by Alexander Blume)
Initialising connection to Enrichr website
library(enrichR)
#> Welcome to enrichR
#> Checking connections ...
#> Enrichr ... Connection is Live!
#> FlyEnrichr ... Connection is Live!
#> WormEnrichr ... Connection is Live!
#> YeastEnrichr ... Connection is Live!
#> FishEnrichr ... Connection is Live!
#> OxEnrichr ... Connection is Live!
listEnrichrSites()
#> Enrichr ... Connection is Live!
#> FlyEnrichr ... Connection is Live!
#> WormEnrichr ... Connection is Live!
#> YeastEnrichr ... Connection is Live!
#> FishEnrichr ... Connection is Live!
#> OxEnrichr ... Connection is Live!
setEnrichrSite("Enrichr") # Human genes
#> Connection changed to https://maayanlab.cloud/Enrichr/
#> Connection is Live!
Select gene-set libraries
List all available databases from Enrichr.
dbs <- listEnrichrDbs()
head(dbs)
| geneCoverage | genesPerTerm | libraryName | numTerms | appyter | categoryId |
|---|---|---|---|---|---|
| 13362 | 275 | Genome_Browser_PWMs | 615 | ea115789fcbf12797fd692cec6df0ab4dbc79c6a | 1 |
| 27884 | 1284 | TRANSFAC_and_JASPAR_PWMs | 326 | 7d42eb43a64a4e3b20d721fc7148f685b53b6b30 | 1 |
| 6002 | 77 | Transcription_Factor_PPIs | 290 | 849f222220618e2599d925b6b51868cf1dab3763 | 1 |
| 47172 | 1370 | ChEA_2013 | 353 | 7ebe772afb55b63b41b79dd8d06ea0fdd9fa2630 | 7 |
| 47107 | 509 | Drug_Perturbations_from_GEO_2014 | 701 | ad270a6876534b7cb063e004289dcd4d3164f342 | 7 |
| 21493 | 3713 | ENCODE_TF_ChIP-seq_2014 | 498 | 497787ebc418d308045efb63b8586f10c526af51 | 7 |
Select the 2023 GO databases.
dbs <- c("GO_Molecular_Function_2023", "GO_Cellular_Component_2023",
"GO_Biological_Process_2023")
Perform analysis
Without background
Query with enrichr() using example genes available from the package.
# Load example input genes
data(input)
length(input)
#> [1] 375
head(input)
#> [1] "Nsun3" "Polrmt" "Nlrx1" "Sfxn5" "Zc3h12c" "Slc25a39"
enriched <- enrichr(input, dbs)
#> Uploading data to Enrichr... Done.
#> Querying GO_Molecular_Function_2023... Done.
#> Querying GO_Cellular_Component_2023... Done.
#> Querying GO_Biological_Process_2023... Done.
#> Parsing results... Done.
Now view the "GO_Biological_Process_2023" results from the enriched object.
head(enriched[["GO_Biological_Process_2023"]])
| Term | Overlap | P.value | Adjusted.P.value | Old.P.value | Old.Adjusted.P.value | Odds.Ratio | Combined.Score | Genes |
|---|---|---|---|---|---|---|---|---|
| Mitochondrial Transcription (GO:0006390) | 3/12 | 0.0012685 | 0.7123925 | 0 | 0 | 17.577061 | 117.23788 | TFAM;POLRMT;TFB1M |
| Alpha-Amino Acid Metabolic Process (GO:1901605) | 4/29 | 0.0019937 | 0.7123925 | 0 | 0 | 8.452830 | 52.55773 | SRR;ALDH6A1;KMO;GNMT |
| Protein Transmembrane Import Into Intracellular Organelle (GO:0044743) | 4/32 | 0.0028882 | 0.7123925 | 0 | 0 | 7.546015 | 44.12249 | DNAJC19;TIMM44;TRIM37;PEX1 |
| Neutrophil Degranulation (GO:0043312) | 2/5 | 0.0033774 | 0.7123925 | 0 | 0 | 35.070599 | 199.57464 | VAMP8;STXBP2 |
| Medium-Chain Fatty Acid Biosynthetic Process (GO:0051792) | 2/5 | 0.0033774 | 0.7123925 | 0 | 0 | 35.070599 | 199.57464 | ABHD3;OXSM |
| Mitochondrial RNA Metabolic Process (GO:0000959) | 3/20 | 0.0058819 | 0.7123925 | 0 | 0 | 9.301708 | 47.77237 | TFAM;POLRMT;TFB1M |
With background
You can now add background genes when using enrichr().
# Load example background
data(background)
length(background)
#> [1] 20625
head(background)
#> [1] "A1BG" "A2M" "NAT1" "NAT2" "SERPINA3" "AADAC"
enriched2 <- enrichr(input, dbs, background = background)
#> Uploading data to Speedrichr...
#> - Your gene set... Done.
#> - Your background... Done.
#> Getting enrichment results...
#> - GO_Molecular_Function_2023... Done.
#> - GO_Cellular_Component_2023... Done.
#> - GO_Biological_Process_2023... Done.
#> Parsing results... Done.
Now view the "GO_Biological_Process_2023" results from the enriched2 object.
head(enriched2[["GO_Biological_Process_2023"]])
| Term | Rank | P.value | Adjusted.P.value | Old.P.value | Old.Adjusted.P.value | Odds.Ratio | Combined.Score | Genes |
|---|---|---|---|---|---|---|---|---|
| Mitochondrial Transcription (GO:0006390) | 1 | 0.0003711 | 0.240515 | 0 | 0 | 27.116000 | 214.19193 | TFAM;POLRMT;TFB1M |
| Alpha-Amino Acid Metabolic Process (GO:1901605) | 2 | 0.0004145 | 0.240515 | 0 | 0 | 13.057671 | 101.69976 | SRR;ALDH6A1;KMO;GNMT |
| Protein Transmembrane Import Into Intracellular Organelle (GO:0044743) | 3 | 0.0006097 | 0.240515 | 0 | 0 | 11.656913 | 86.29136 | DNAJC19;TIMM44;TRIM37;PEX1 |
| Monocarboxylic Acid Biosynthetic Process (GO:0072330) | 4 | 0.0012176 | 0.240515 | 0 | 0 | 6.816532 | 45.74506 | ALDH1A3;SRR;SCP2;OXSM;MCAT |
| Neutrophil Degranulation (GO:0043312) | 5 | 0.0014663 | 0.240515 | 0 | 0 | 54.031872 | 352.55862 | VAMP8;STXBP2 |
| Medium-Chain Fatty Acid Biosynthetic Process (GO:0051792) | 6 | 0.0014663 | 0.240515 | 0 | 0 | 54.031872 | 352.55862 | ABHD3;OXSM |
By default, the results table from analysis with a background does not have the ‘Overlap’ column. We can calculate the annotated genes in each term from GMT files and replace the ‘Rank’ column with ‘Overlap’ by setting include_overlap = TRUE.
enriched3 <- enrichr(input, dbs, background = background, include_overlap = TRUE)
#> Uploading data to Speedrichr...
#> - Your gene set... Done.
#> - Your background... Done.
#> Getting enrichment results...
#> - GO_Molecular_Function_2023... Done.
#> - Download GMT file... Done.
#> - GO_Cellular_Component_2023... Done.
#> - Download GMT file... Done.
#> - GO_Biological_Process_2023... Done.
#> - Download GMT file... Done.
#> Parsing results... Done.
Now view the "GO_Biological_Process_2023" results from the enriched3 object.
head(enriched3[["GO_Biological_Process_2023"]])
| Term | Overlap | P.value | Adjusted.P.value | Old.P.value | Old.Adjusted.P.value | Odds.Ratio | Combined.Score | Genes |
|---|---|---|---|---|---|---|---|---|
| Mitochondrial Transcription (GO:0006390) | 3/12 | 0.0003711 | 0.240515 | 0 | 0 | 27.116000 | 214.19193 | TFAM;POLRMT;TFB1M |
| Alpha-Amino Acid Metabolic Process (GO:1901605) | 4/29 | 0.0004145 | 0.240515 | 0 | 0 | 13.057671 | 101.69976 | SRR;ALDH6A1;KMO;GNMT |
| Protein Transmembrane Import Into Intracellular Organelle (GO:0044743) | 4/32 | 0.0006097 | 0.240515 | 0 | 0 | 11.656913 | 86.29136 | DNAJC19;TIMM44;TRIM37;PEX1 |
| Monocarboxylic Acid Biosynthetic Process (GO:0072330) | 5/65 | 0.0012176 | 0.240515 | 0 | 0 | 6.816532 | 45.74506 | ALDH1A3;SRR;SCP2;OXSM;MCAT |
| Neutrophil Degranulation (GO:0043312) | 2/5 | 0.0014663 | 0.240515 | 0 | 0 | 54.031872 | 352.55862 | VAMP8;STXBP2 |
| Medium-Chain Fatty Acid Biosynthetic Process (GO:0051792) | 2/5 | 0.0014663 | 0.240515 | 0 | 0 | 54.031872 | 352.55862 | ABHD3;OXSM |
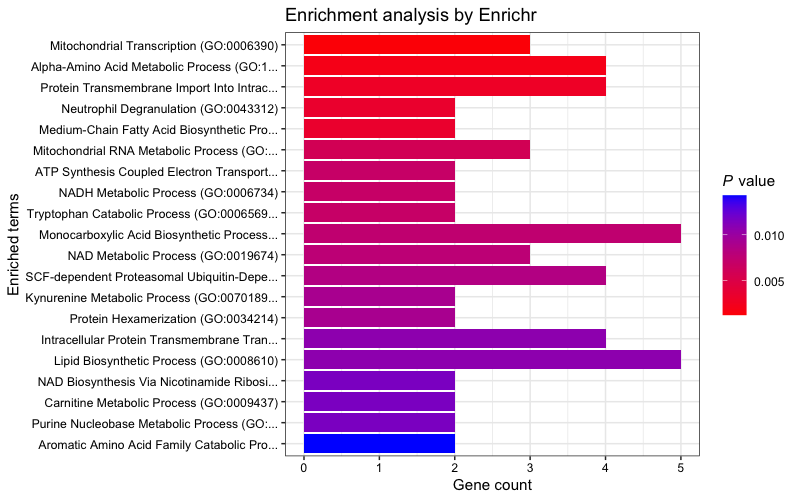
Visualise results
Plot the "GO_Biological_Process_2023" results. (Plotting function contributed by I-Hsuan Lin)
plotEnrich(enriched[["GO_Biological_Process_2023"]], showTerms = 20, numChar = 40,
y = "Count", orderBy = "P.value")
Export results
Export Enrichr results as text or Excel files. By default (i.e. outFile = "txt"), the results from all the selected databases are saved into individual text files. When using outFile = "excel", the results are saved into worksheets in a single Excel 2007 (XLSX) file. (Print function contributed by I-Hsuan Lin and Kai Hu)
# To text files
printEnrich(enriched)
# To Excel
printEnrich(enriched, outFile = "excel")
Using enrichR behind a proxy
If your computer is behind an HTTP or HTTPS proxy, you can set the RCurl Proxy options explicitly using RCurlOptions and enrichR will use the provided settings to connect to the Enrichr database via httr::use_proxy().
For example:
options(RCurlOptions = list(proxy = 'http://ip_or_url',
proxyusername = 'myuser',
proxypassword = 'mypwd',
proxyport = 'port_num',
proxyauth = 'basic'))
References
Kuleshov, Maxim V., Matthew R. Jones, Andrew D. Rouillard, Nicolas F. Fernandez, Qiaonan Duan, Zichen Wang, Simon Koplev, et al. 2016. “Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update.” Nucleic Acids Res 44 (Web Server issue): W90–97. https://doi.org/10.1093/nar/gkw377.