Description
Exact Post Selection Inference with Applications to the Lasso.
Description
Implements the conditional estimation procedure of Lee, Sun, Sun and Taylor (2016) <doi:10.1214/15-AOS1371>. This procedure allows hypothesis testing on the mean of a normal random vector subject to linear constraints.
README.md
epsiwal
Implements conditional inference on normal variates as described in Lee, Sun, Sun and Taylor, "Exact Post Selection Inference, with Application to the Lasso."
-- Steven E. Pav, [email protected]
Installation
This package may be installed from CRAN; the latest version may be found on github via devtools, or installed via drat:
# CRAN
install.packages(c("epsiwal"))
# devtools
if (require(devtools)) {
# latest greatest
install_github("shabbychef/epsiwal")
}
# via drat:
if (require(drat)) {
drat:::add("shabbychef")
install.packages("epsiwal")
}
Basic Usage
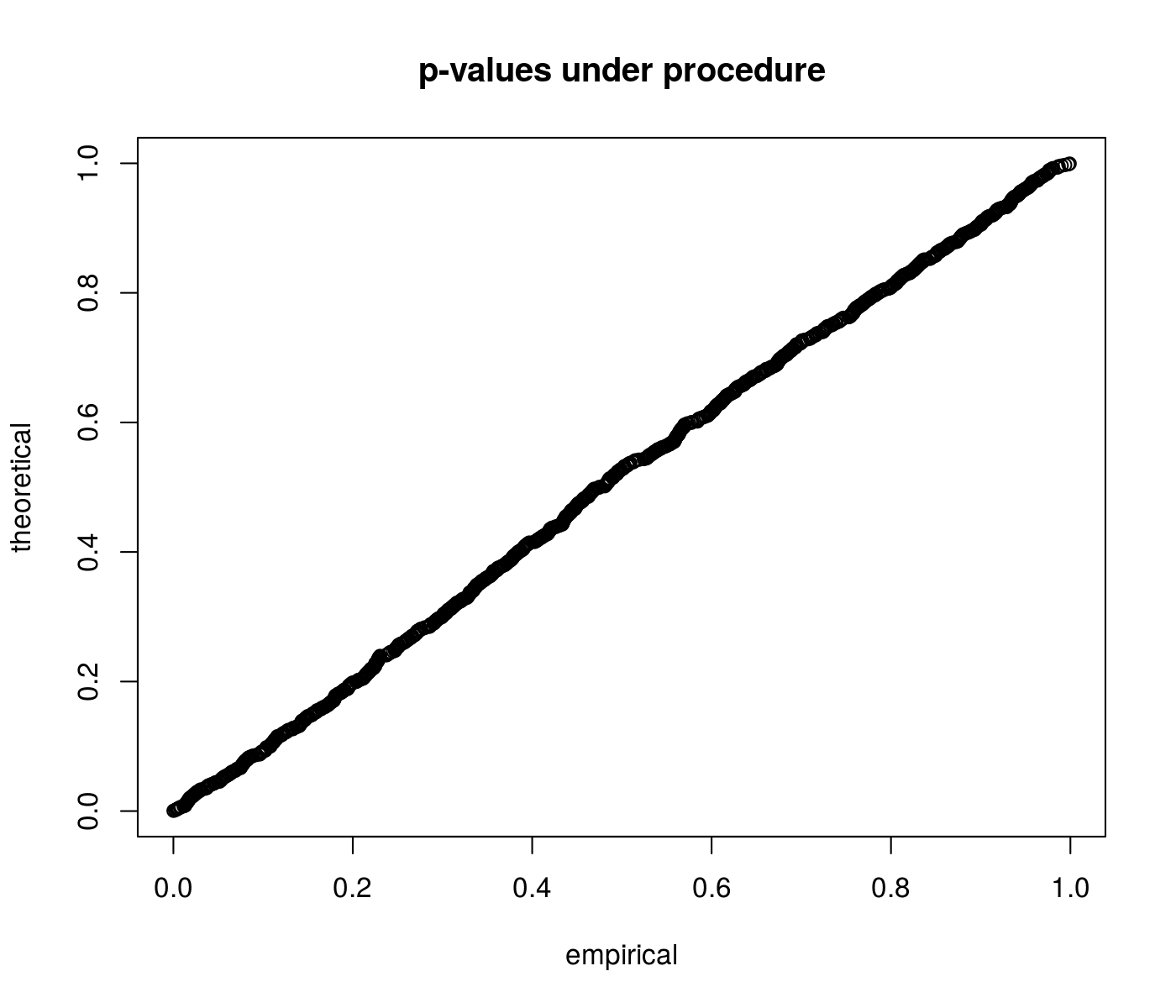
First we perform some simulations under the null to show that the p-values are uniform. We draw a normal vector with identity covariance and zero mean, then flip the sign of each element to make them positive. We then perform inference on the sum of the mean values.
library(epsiwal)
p <- 20
mu <- rep(0, p)
Sigma <- diag(p)
A <- -diag(p)
b <- rep(0, p)
eta <- rep(1, p)
Sigma_eta <- diag(Sigma)
eta_mu <- as.numeric(t(eta) %*% mu)
set.seed(1234)
pvals <- replicate(1000, {
y <- rnorm(p, mean = mu, sd = sqrt(diag(Sigma)))
ay <- abs(y)
pconnorm(y = ay, A = A, b = b, eta = eta, Sigma_eta = Sigma_eta,
eta_mu = eta_mu)
})
qqplot(pvals, qunif(ppoints(length(pvals))), main = "p-values under procedure",
ylab = "theoretical", xlab = "empirical")
library(epsiwal)
p <- 20
mu <- rep(0, p)
Sigma <- diag(p)
A <- -diag(p)
b <- rep(0, p)
eta <- rep(1, p)
Sigma_eta <- diag(Sigma)
eta_mu <- as.numeric(t(eta) %*% mu)
type_I <- 0.05
set.seed(1234)
civals <- replicate(5000, {
y <- rnorm(p, mean = mu, sd = sqrt(diag(Sigma)))
ay <- abs(y)
ci <- ci_connorm(y = ay, A = A, b = b, p = type_I,
eta = eta, Sigma_eta = Sigma_eta)
})
cat("Empirical coverage of the", type_I, "confidence bound is around",
mean(civals < eta_mu), ".\n")
## Empirical coverage of the 0.05 confidence bound is around 0.052 .
See also
- The original paper, by Lee, J. D., Sun, D. L., Sun, Y. and Taylor, J. E. Exact post-selection inference, with application to the Lasso.
- The
PSATpackage, which supports similar procedures, but is not yet on CRAN. - The
SelectiveInferencepackage, which implements similar inferential procedures under quadratic constraints, as detailed in Tibshirani, R. J., Taylor, J., Lockhart, R. and Tibshirani, R. Exact Post-Selection Inference for Sequential Regression Procedures.
