Preprocessing Experience Sampling Method (ESM) Data.
esmtools
This R package proposes tools for preprocessing and analyzing data collected through Ecological Momentary Assessment (EMA) or Experience Sampling Method (ESM) studies. The ‘esmtools’ package provides a range of functions to support preprocessing, to help researchers gain valuable insights from their datasets, and to help report their preprocessing practices and the quality of the preprocessing of the dataset.
It has been developed as part of a set of tools introduced by Revol and al. (in preparation). Among developed tools, you can find the ESM preprocessing Gallery (https://preprocess.esmtools.com/) which provides instructions, R code, and practical examples to preprocess your ESM dataset following a 5 steps framework.
Please remember to cite us if you find our package, framework, and resources helpful in your study:
- Revol, J., Carlier, C., Lafit, G., Verhees, M., Sels, L., & Ceulemans, E. (in preparation). Preprocessing ESM data: A step-by-step framework, reporting templates, and R code website
Installation
You can install the package like so:
install.packages("esmtools")
Once installed, you can load the package, as follows:
library(esmtools)
To learn more about the available functions and their usage, please refer to the documentation: https://package.esmtools.com/.
Plot example
The calendar plot can provide a comprehensive overview of the time the data was collected, allowing you to identify periods (e.g., holidays vs. regular weeks), or detect missing observations for the whole dataset or a specific participant.
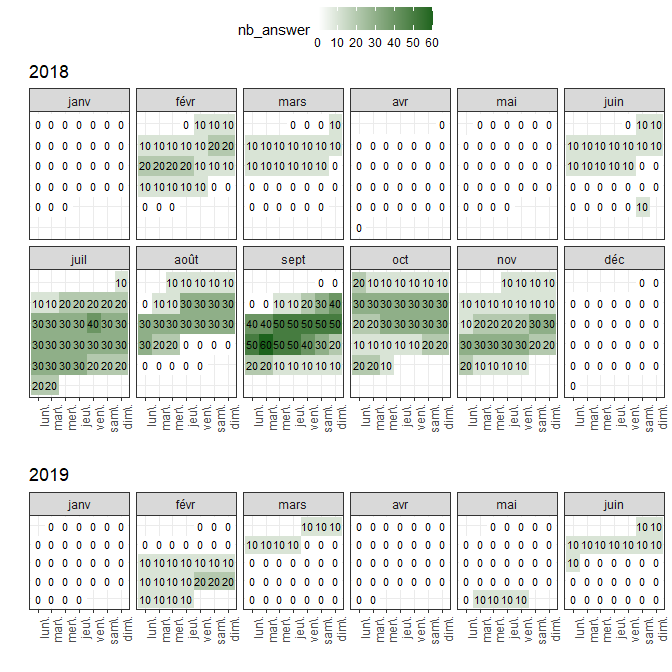
Templates
‘esmtools’ provides a set of RMarkdown templates that are bundled with the package. The templates are designed to assist in the reporting of the preprocessing phase and the assessment of data quality, allowing researchers to document and share their data preprocessing workflows efficiently. These templates can be accessed using the ‘esmtools::use_template()’ function, which copies the template files to your working directory. Available templates are: “preprocess_report”, “advanced_preprocess_report”, “data_quality_report”. For instance:
esmtools::use_template("preprocess_report")
For further information, please refer to the Preprocessing report and the Data characteristics report topics of the Preprocessing ESM Gallery.
Reporting tool examples
Among the functions of this package, you can find functions that support reporting the preprocessing steps. For instance, you can highlight part of the text using the ‘txt()’ function:
Modification 1: The value of the x variable has been changed to …
Additionally, the ‘button()’ and ‘endbutton()’ functions from the ‘esmtools’ package delimite part of the document that can be revealed using a button. It is particularly useful to hide non-essential elements (but still important to report) to improve readibility.
print(1 + 1)
#> [1] 2
print(2 + 2)
#> [1] 4
Datasets examples
The package contains three dataset examples. The first one is a simulated dataset (‘esmdata_sim’) that is used in the ESM Preprocessing Gallery website (for further details see https://preprocess.esmtools.com/terminology.html). The second dataset (‘esmdata_raw’) is used as an illustrative dataset in the article (further details in the article). Finally, the third dataset (‘esmdata_preprocessed’), is the output of the preprocessing applied to the raw dataset. The preprocessing that was done can be found in the associated preprocessing report (for further details see the report).
There are two ways to import the datasets:
- Using the name of the dataset.
library(esmtools)
data = esmdata_raw
- Importing the csv version by reconstructing the path to the file (stored in the package).
file_path = system.file("extdata", "esmdata_raw.csv", package="esmtools")
data = read.csv(file_path)
About
To cite esmtools, please use:
- Revol, J., Carlier, C., Lafit, G., Verhees, M., Sels, L., & Ceulemans, E. (2023). Preprocessing ESM data: a step-by-step framework, tutorial website, R package, and reporting templates
For any questions, bug reports, or suggestions, please reach out to our team:
We value your feedback and are committed to improving ‘esmtools’ based on your needs and requirements.
Licence
The package source code in this repository is licensed under the GNU General Public License, version 3 license. The documentation, vignettes, and other website materials by Jordan Revol are licensed under CC BY 4.0.