Explore the Exact Bootstrap Method.
exactamente
‘exactamente’ is an R package that offers a collection of tools to assist researchers and data analysts in exploring bootstrap methods on small sample size data. Bootstrap methods are widely used for estimating the sampling distribution of an estimator by resampling with replacement from the original sample.
This package is focused particularly on the exact bootstrap as described in Kisielinska (2013). This method is advantageous for bootstrapping small data sets where standard methods might be inadequate.
For a given sample size N, the exact bootstrap generates all N^N resamples, including permutations such as [5, 5, 3], [5, 3, 5], and [3, 5, 5] as distinct resamples.
Furthermore, ‘exactamente’ provides a standard bootstrap function where the user can specify the desired number of resamples, allowing for direct comparison between the exact method and conventional bootstrap techniques.
Here is the the step-by-step process that the ‘exactamente’ package functions use to perform their bootstrap resampling and compute the summary statistics and density estimates:
- Bootstrap Resampling: Each bootstrap method starts by generating a collection of bootstrap samples, also known as resamples.
- For exact_bootstrap(), the function generates N^N resamples, where each different permutation is treated as a unique resample.
- For reg_bootstrap(), the function generates a user-specified number of resamples by sampling with replacement from the original sample.
Compute the Resample Statistics: For each resample, the function computes a statistic of interest, such as the mean or the median. The function uses the user-specified ‘anon’ function for this computation. The ‘anon’ function defaults to the mean if not specified by the user.
Process Bootstrap Statistics: This is a common process for both methods. After generating the resample statistics, each function calls process_bootstrap_stats() to derive the summary statistics and density estimates.
Density Estimation: In process_bootstrap_stats(), it first calculates the kernel density estimate of the bootstrap statistics using the stats::density() function. If user-specified ‘density_args’ are provided, those are passed to the density function. The density estimate provides a smoothed representation of the distribution of the bootstrap statistics.
Summary Statistics: After generating the density estimate, process_bootstrap_stats() computes various summary statistics for the bootstrap statistics:
- The total number of resamples (nres).
- The mode, defined as the value with the highest frequency in the bootstrap statistics.
- The median, defined as the middle value when the bootstrap statistics are sorted.
- The mean, defined as the average of the bootstrap statistics.
- The standard deviation, which measures the dispersion of the bootstrap statistics.
- The lower and upper bootstrap confidence interval (lCI and uCI), defined by the user-specified percentiles (‘lb’ and ‘ub’). The default is the 2.5th percentile and the 97.5th percentile, giving a 95% confidence interval.
- Return Result: Lastly, process_bootstrap_stats() returns a list containing the density estimate and the summary statistics. The bootstrap function then assigns a specific class to the result (‘extboot’, or ‘regboot’) and returns the result to the user.
By providing these detailed outputs, the ‘exactamente’ package enables users to thoroughly investigate the characteristics of the bootstrap distribution and the behavior of the bootstrap estimator under different resampling methods.
Installation
You can install the released version of ‘exactamente’ from CRAN:
install.packages("exactamente")
You can install the development version of ‘exactamente’ from GitHub like so:
# install.packages("devtools")
devtools::install_github("mightymetrika/exactamente")
Example Usage
After installation, you can load the ‘exactamente’ package using the library function.
library(exactamente)
To utilize the fundamental tools provided by ‘exactamente’, begin by creating a numeric vector of data. You can then use the _bootstrap functions to obtain objects holding summary statistics and density estimates of the respective bootstrap distributions. In the following example, we use the [1, 2, 7] sample vector and take the mean as our test statistic.
data <- c(1, 2, 7)
# Run exact bootstrap
e_res <- exact_bootstrap(data)
# Run regular bootstrap
set.seed(183)
r_res <- reg_bootstrap(data)
Both _bootstrap functions comes with plot() and summary() methods for further investigation of the bootstrap results.
res <- list(e_res, r_res)
lapply(res, plot)
#> [[1]]
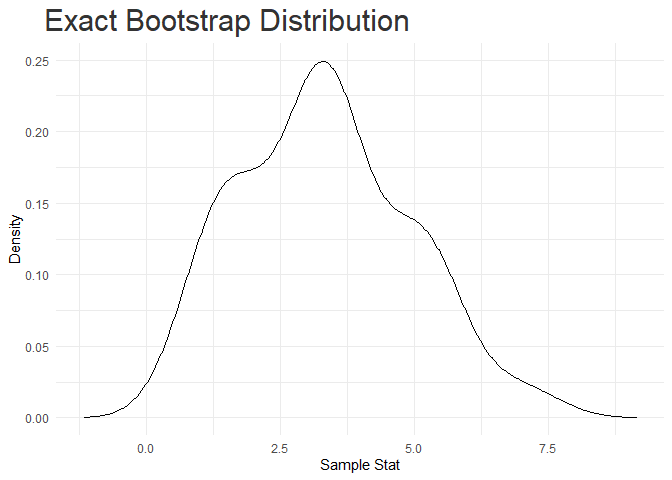
#>
#> [[2]]
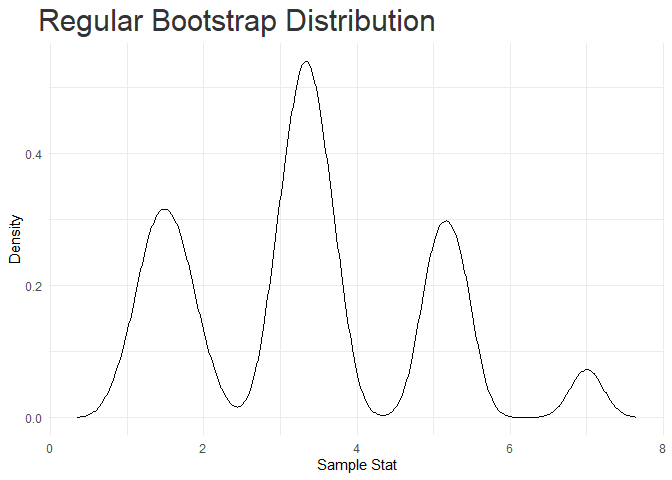
lapply(res, summary)
#> [[1]]
#> Method nres mode median mean sd lCI uCI
#> 1 exact_bootstrap 27 3.303687 3.333333 3.333333 1.54422 1.216667 5.916667
#>
#> [[2]]
#> Method nres mode median mean sd lCI uCI
#> 1 reg_bootstrap 10000 3.335788 3.333333 3.3368 1.518024 1 7
‘exactamente’ also includes the e_vs_r() function, which enables direct comparison between the two methods using the same data set.
set.seed(183)
comp_res <- e_vs_r(data)
comp_res$comp_plot
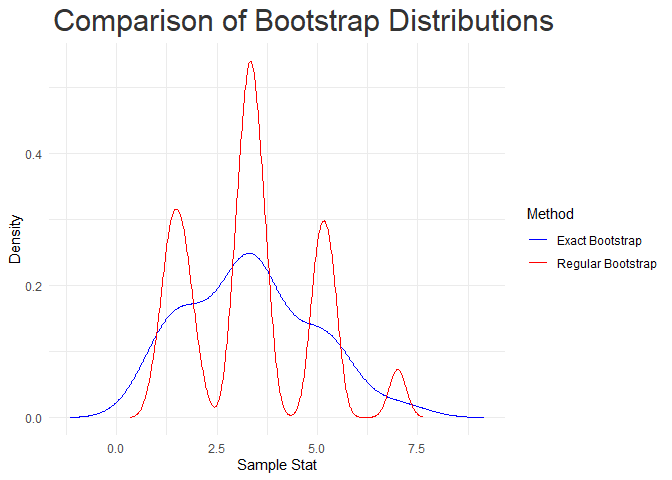
comp_res$summary_table
#> Method nres mode median mean sd lCI uCI
#> 1 exact_bootstrap 27 3.303687 3.333333 3.333333 1.544220 1.216667 5.916667
#> 2 reg_bootstrap 10000 3.335788 3.333333 3.336800 1.518024 1.000000 7.000000
Interactive Shiny App
One of the exciting features of the ‘exactamente’ package is the inclusion of an interactive Shiny app. The app allows you to visually explore and compare the bootstrap methods. It is designed with a user-friendly interface and offers real-time results visualization.
You can access this app with the following command:
exactamente_app()
References
Kisielinska, J. (2013). The exact bootstrap method shown on the example of the mean and variance estimation. Computational Statistics, 28, 1061–1077.