Interactive Interfaces for Results Exploration.
explor 
explor is an R package to allow interactive exploration of multivariate analysis results.
For now on, it is usable the following types of analyses :
| Analysis | Function | Package | Notes |
|---|---|---|---|
| Principal Component Analysis | PCA | FactoMineR | - |
| Correspondance Analysis | CA | FactoMineR | - |
| Multiple Correspondence Analysis | MCA | FactoMineR | - |
| Principal Component Analysis | dudi.pca | ade4 | Qualitative supplementary variables are ignored |
| Correspondance Analysis | dudi.coa | ade4 | - |
| Multiple Correspondence Analysis | dudi.acm | ade4 | Quantitative supplementary variables are ignored |
| Specific Multiple Correspondance Analysis | speMCA | GDAtools | - |
| Multiple Correspondance Analysis | mca | MASS | Quantitative supplementary variables are not supported |
| Principal Component Analysis | princomp | stats | Supplementary variables are ignored |
| Principal Component Analysis | prcomp | stats | Supplementary variables are ignored |
| Correspondance Analysis | textmodel_ca | quanteda.textmodels | Only coordinates are available |
Features
For each type of analysis, explor launches a shiny interactive Web interface which is displayed inside RStudio or in your system Web browser. This interface provides both numerical results as dynamic tables (sortable and searchable thanks to the DT package) and interactive graphics thanks to the scatterD3 package. You can zoom, drag labels, hover points to display tooltips, hover legend items to highlights points, and the graphics are fully updatable with animations which can give some visual clues. You can also export the current plot as an SVG file or get the R code to reproduce it later in a script or document.
Here is a preview of what you will get. Note that the interface is available both in english and french, depending on your locale :
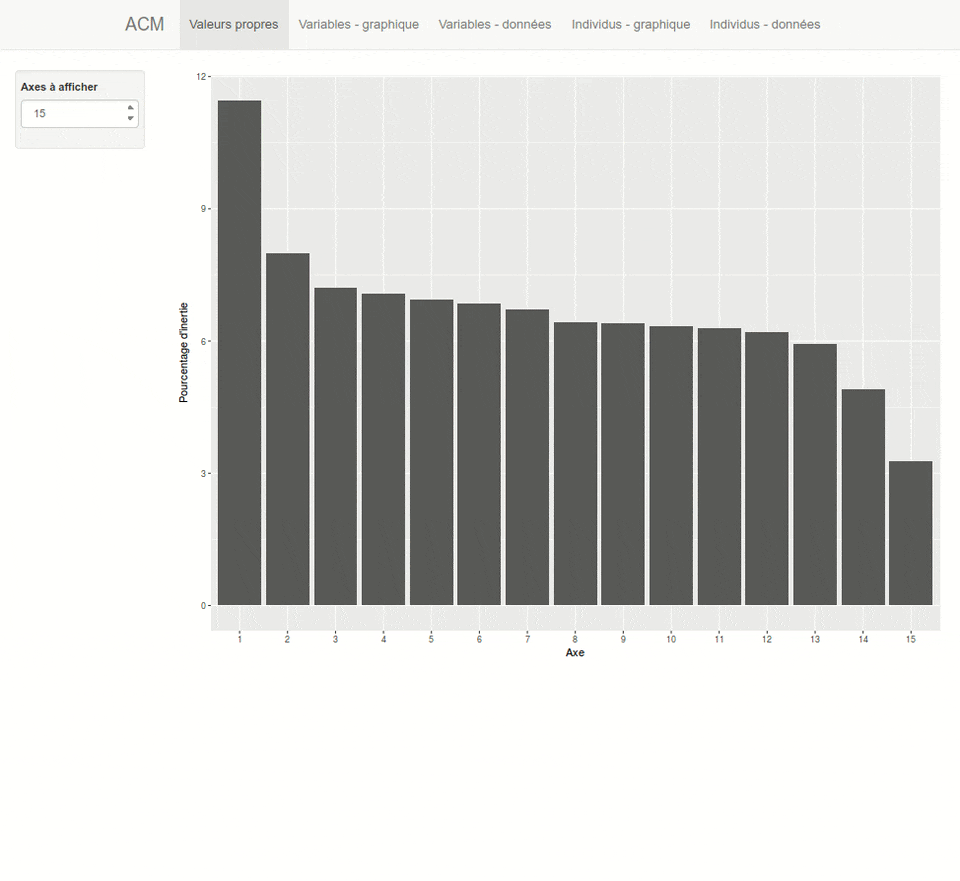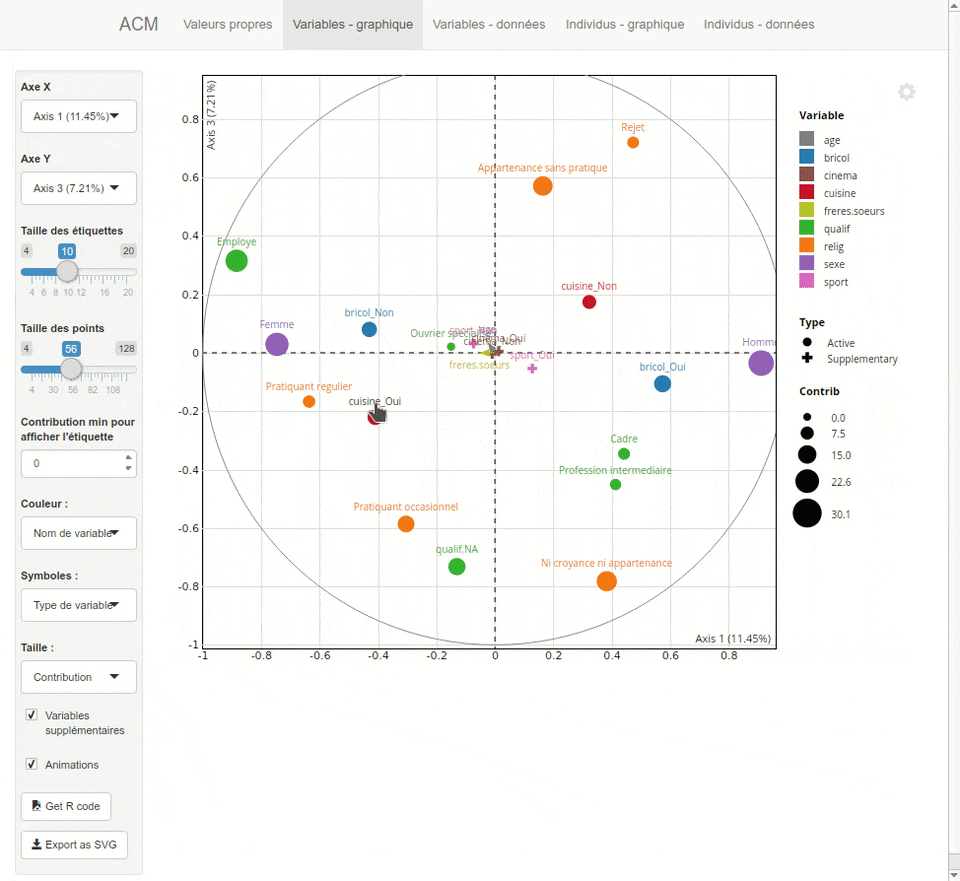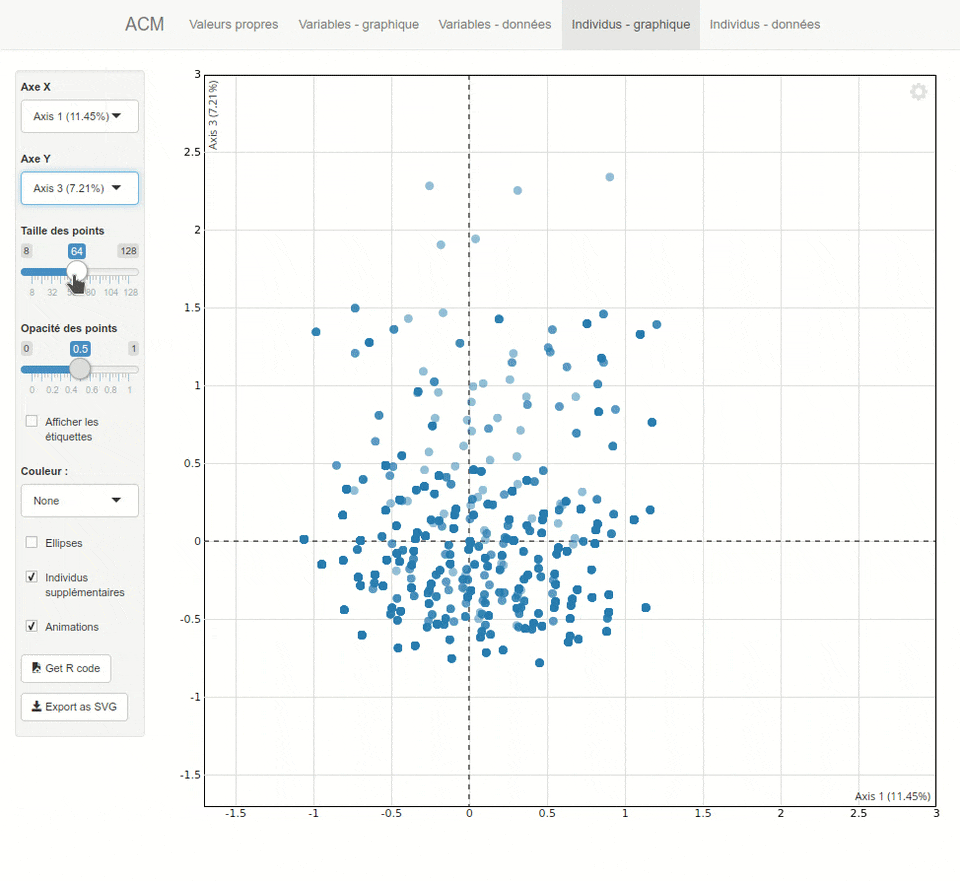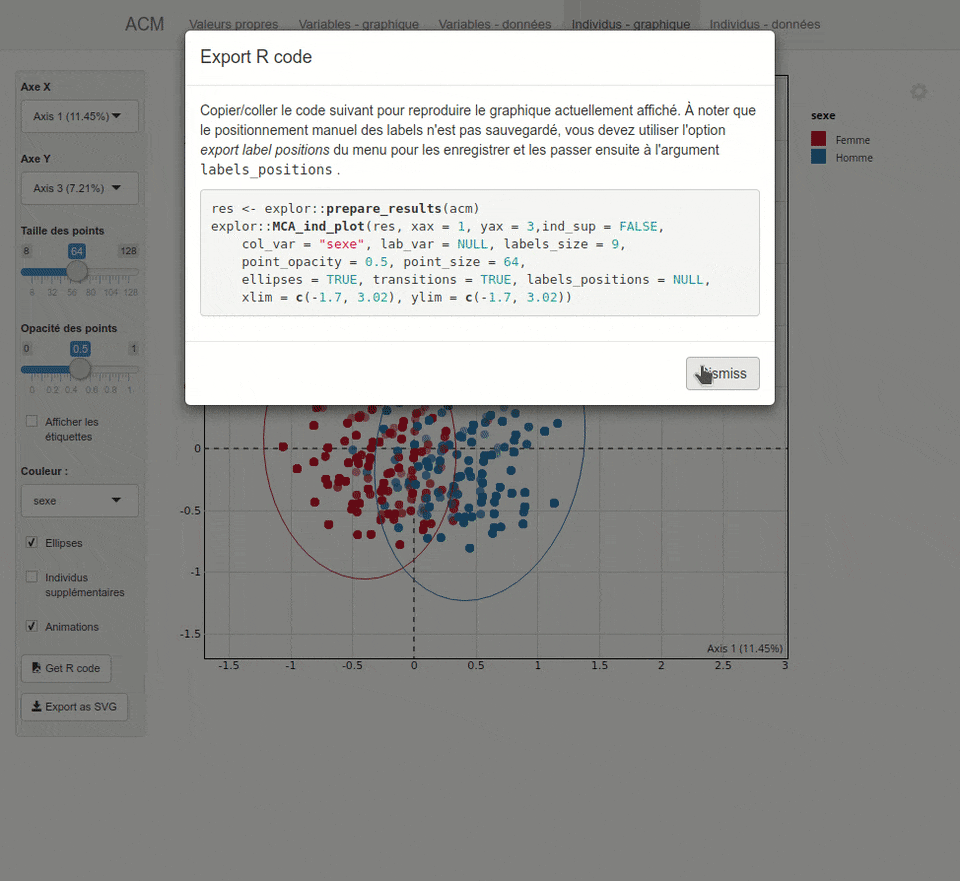
Installation
To get the stable version from CRAN :
install.packages("explor")
To install the latest dev version from GitHub :
install.packages("remotes") # If necessary
remotes::install_github("juba/scatterD3")
remotes::install_github("juba/explor")
Usage
Usage is very simple : you just apply the explor function to the result of one of the supported analysis functions.
Example with a principal correspondence analysis from FactoMineR::PCA :
library(FactoMineR)
library(explor)
data(decathlon)
pca <- PCA(decathlon[,1:12], quanti.sup = 11:12, graph = FALSE)
explor(pca)
Example with a multiple correspondence analysis from FactoMineR::MCA:
data(hobbies)
mca <- MCA(hobbies[1:1000,c(1:8,21:23)],quali.sup = 9:10, quanti.sup = 11, ind.sup = 1:100)
explor(mca)
Documentation and localization
Two vignettes are provided for more detailed documentation :
Depending on your system locale settings, the interface is displayed either in english or in french (other languages can be easily added).