Flexible Tool for Bias Detection, Visualization, and Mitigation.
fairmodels 
Overview
Flexible tool for bias detection, visualization, and mitigation. Use models explained with DALEX and calculate fairness classification metrics based on confusion matrices using fairness_check() or try newly developed module for regression models using fairness_check_regression(). R package fairmodels allows to compare and gain information about various machine learning models. Mitigate bias with various pre-processing and post-processing techniques. Make sure your models are classifying protected groups similarly.
Preview
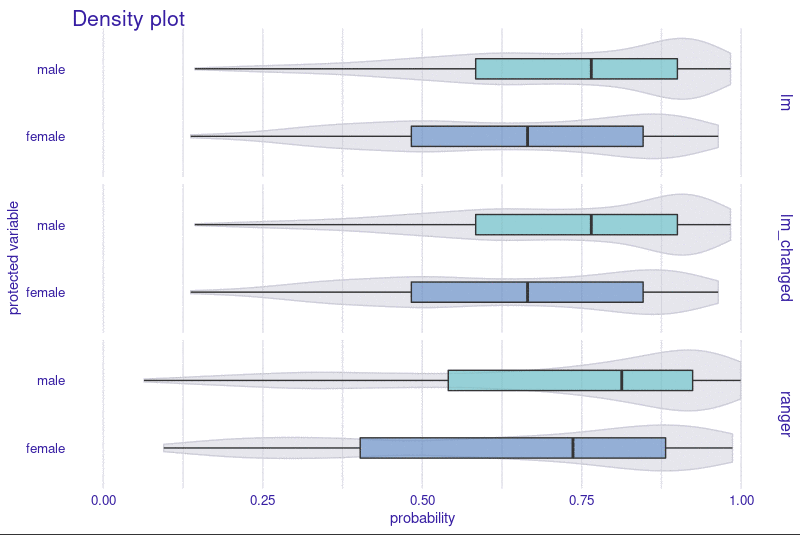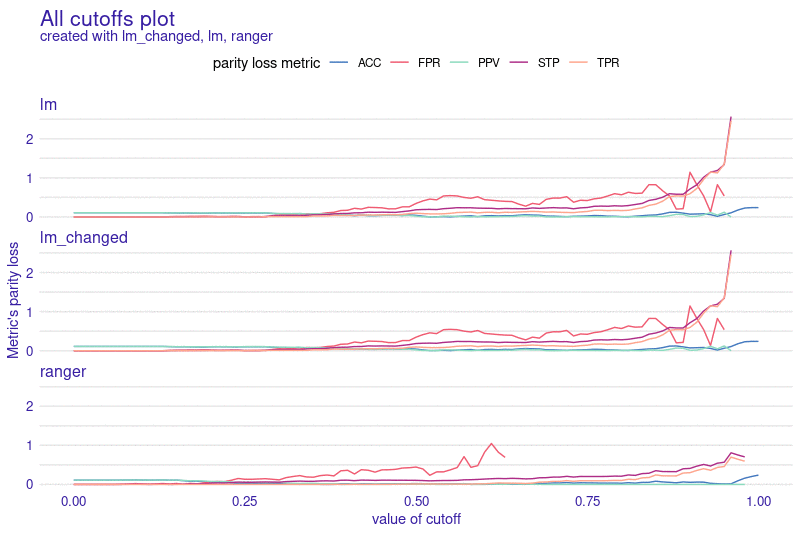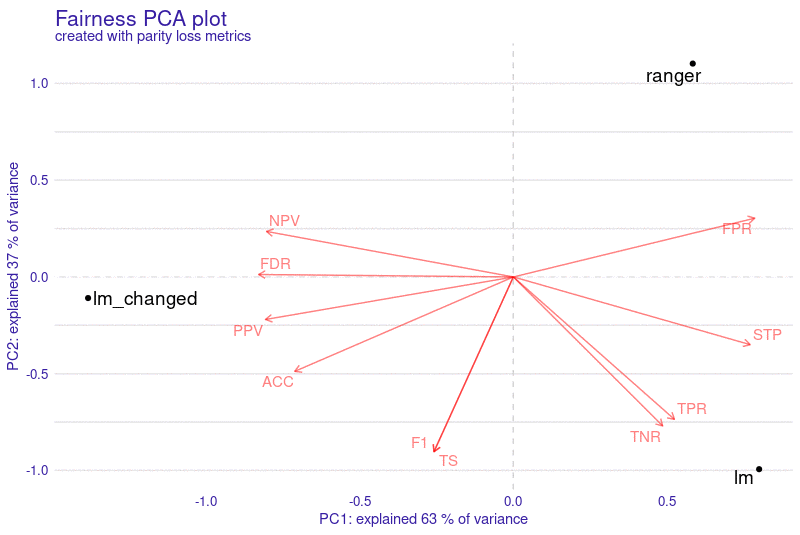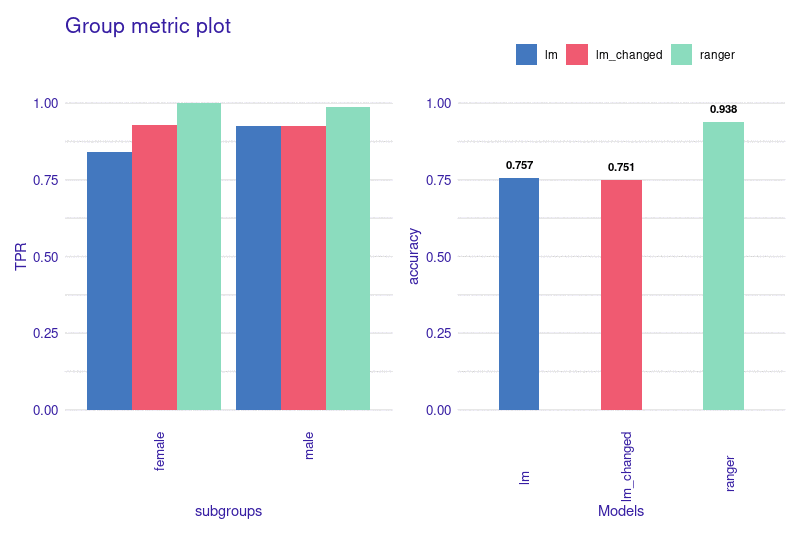
Installation
Install it from CRAN:
install.packages("fairmodels")
or developer version from GitHub:
devtools::install_github("ModelOriented/fairmodels")
Example
Checking fairness is easy!
library(fairmodels)
library(ranger)
library(DALEX)
data("german")
# ------------ step 1 - create model(s) -----------------
lm_model <- glm(Risk~.,
data = german,
family=binomial(link="logit"))
rf_model <- ranger(Risk ~.,
data = german,
probability = TRUE,
num.trees = 200)
# ------------ step 2 - create explainer(s) ------------
# numeric y for explain function
y_numeric <- as.numeric(german$Risk) -1
explainer_lm <- explain(lm_model, data = german[,-1], y = y_numeric)
explainer_rf <- explain(rf_model, data = german[,-1], y = y_numeric)
# ------------ step 3 - fairness check -----------------
fobject <- fairness_check(explainer_lm, explainer_rf,
protected = german$Sex,
privileged = "male")
print(fobject)
plot(fobject)
Compas recidivism data use case: Basic tutorial
Bias mitigation techniques on Adult data: Advanced tutorial
How to evaluate fairness in classification models?
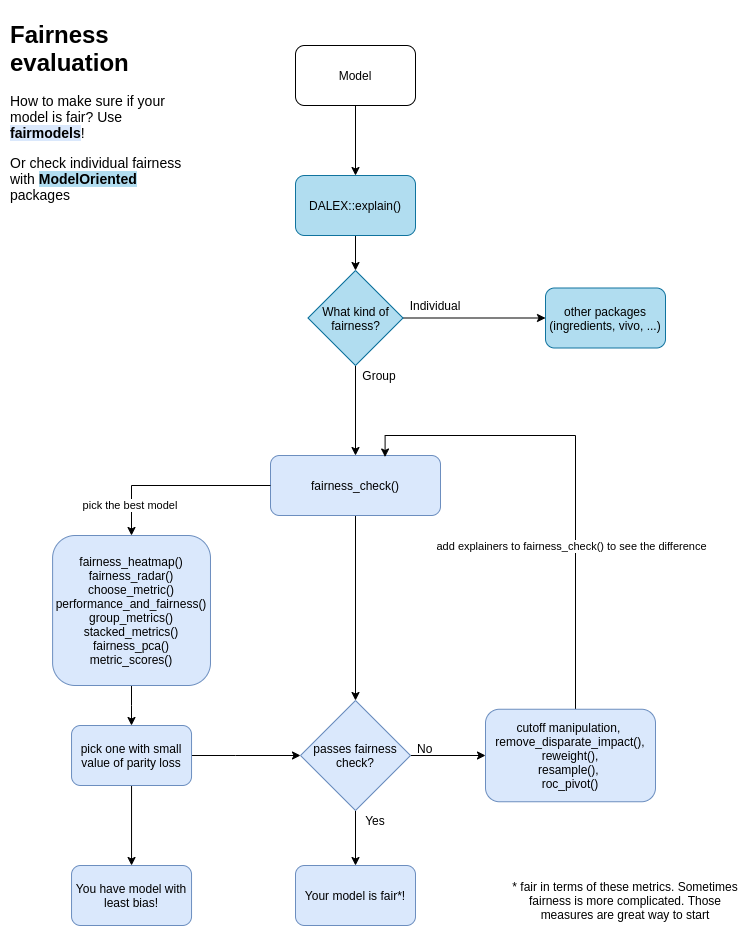
Fairness checking is flexible
fairness_check parameters are
- x, ... -
explainersandfairness_objects(products of fairness_check). - protected - factor with different subgroups as levels. Usually specific race, sex etc...
- privileged - subgroup, base on which to calculate parity loss metrics.
- cutoff - custom cutoff, might be single value - cutoff same for all subgroups or vector - for each subgroup individually. Affecting only explainers.
- label - character vector for every explainer.
Models might be trained on different data, even without protected variable. May have different cutoffs which gives different values of metrics. fairness_check() is place where explainers and fairness_objects are checked for compatibility and then glued together.
So it is possible to to something like this:
fairness_object <- fairness_check(explainer1, explainer2, ...)
fairness_object <- fairness_check(explainer3, explainer4, fairness_object, ...)
even with more fairness_objects!
If one is even more keen to know how fairmodels works and what are relations between objects, please look at this diagram class diagram
Metrics used
There are 12 metrics based on confusion matrix :
| Metric | Formula | Full name | fairness names while checking among subgroups |
|---|---|---|---|
| TPR | 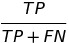 | true positive rate | equal opportunity |
| TNR | 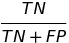 | true negative rate | |
| PPV | 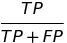 | positive predictive value | predictive parity |
| NPV | 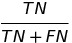 | negative predictive value | |
| FNR | 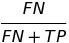 | false negative rate | |
| FPR | 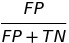 | false positive rate | predictive equality |
| FDR | 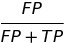 | false discovery rate | |
| FOR | 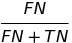 | false omission rate | |
| TS | 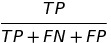 | threat score | |
| STP | 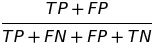 | statistical parity | statistical parity |
| ACC | 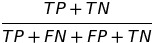 | accuracy | Overall accuracy equality |
| F1 |  | F1 score |
and their parity loss.
How is parity loss calculated?
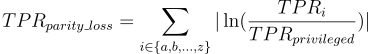
Where i denotes the membership to unique subgroup from protected variable. Unprivileged subgroups are represented by small letters and privileged by simply "privileged".
some fairness metrics like Equalized odds are satisfied if parity loss in both TPR and FPR is low
How easy it is to add custom fairness metric?
It is relatively easy! Check it out here
Fairness in regression
R package fairmodels has support for regression models. Check fairness using fairness_check_regression() to approximate classification fairness metrics in regression setting. Plot object with plot() to visualize fairness check or with plot_density() to see model's output.
Related works
Zafar, Valera, Rodriguez, Gummadi (2017) https://arxiv.org/pdf/1610.08452.pdf
Barocas, Hardt, Narayanan (2019) https://fairmlbook.org/
Steinberg, Daniel & Reid, Alistair & O'Callaghan, Simon. (2020). Fairness Measures for Regression via Probabilistic Classification. - https://arxiv.org/pdf/2001.06089.pdf.
