Fast and Stable Fitting of Generalized Linear Models using 'RcppEigen'.
Overview of ‘fastglm’
The ‘fastglm’ package is a re-write of glm() using RcppEigen designed to be computationally efficient and algorithmically stable.
Installing the ‘fastglm’ package
Install the development version using the devtools package:
devtools::install_github("jaredhuling/fastglm")
or by cloning and building using R CMD INSTALL
Quick Usage Overview
Load the package:
library(fastglm)
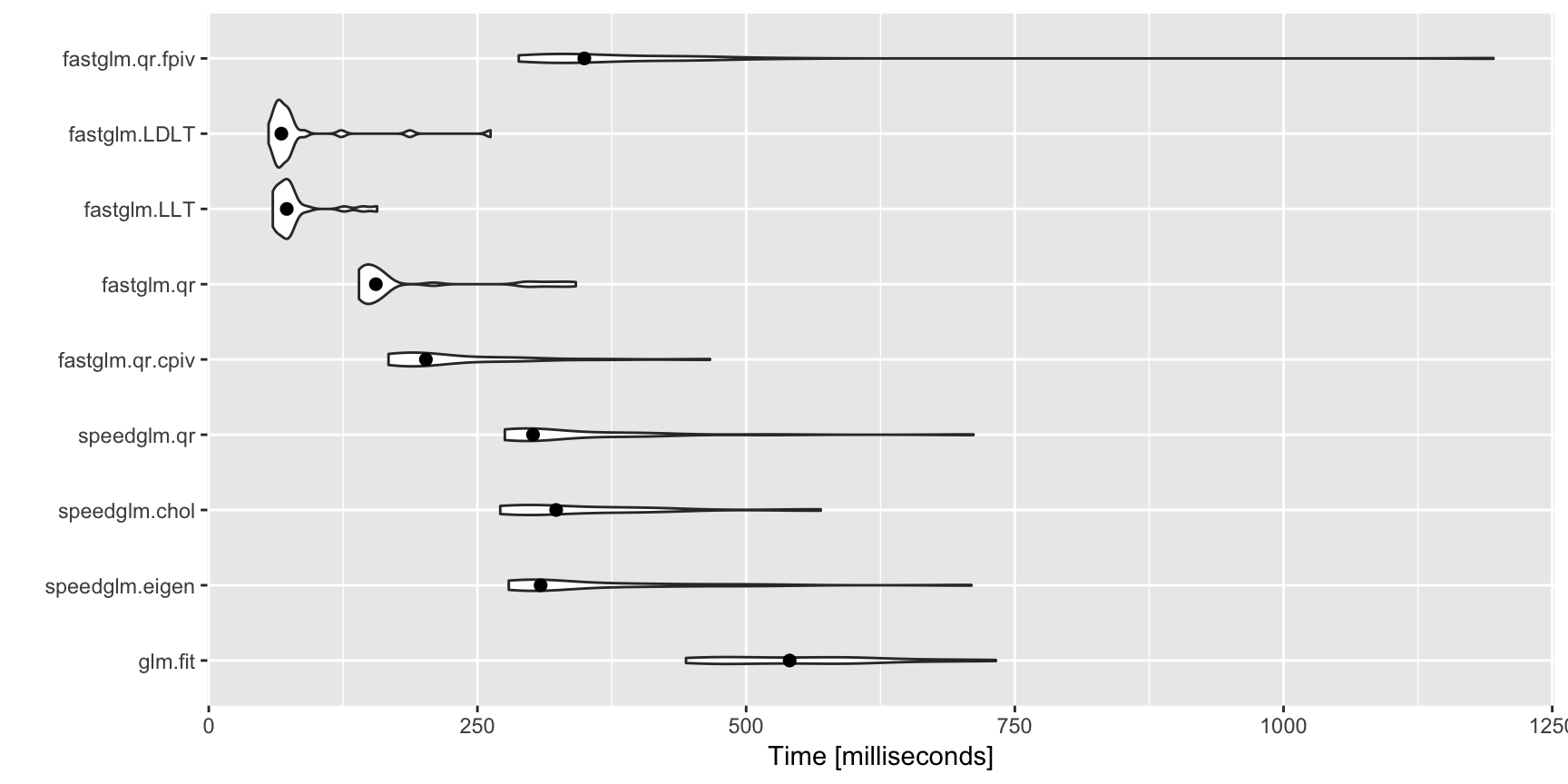
A (not comprehensive) comparison with glm.fit() and speedglm.wfit():
library(speedglm)
library(microbenchmark)
library(ggplot2)
set.seed(123)
n.obs <- 10000
n.vars <- 100
x <- matrix(rnorm(n.obs * n.vars, sd = 3), n.obs, n.vars)
Sigma <- 0.99 ^ abs(outer(1:n.vars, 1:n.vars, FUN = "-"))
x <- MASS::mvrnorm(n.obs, mu = runif(n.vars, min = -1), Sigma = Sigma)
y <- 1 * ( drop(x[,1:25] %*% runif(25, min = -0.1, max = 0.10)) > rnorm(n.obs))
ct <- microbenchmark(
glm.fit = {gl1 <- glm.fit(x, y, family = binomial())},
speedglm.eigen = {sg1 <- speedglm.wfit(y, x, intercept = FALSE,
family = binomial())},
speedglm.chol = {sg2 <- speedglm.wfit(y, x, intercept = FALSE,
family = binomial(), method = "Chol")},
speedglm.qr = {sg3 <- speedglm.wfit(y, x, intercept = FALSE,
family = binomial(), method = "qr")},
fastglm.qr.cpiv = {gf1 <- fastglm(x, y, family = binomial())},
fastglm.qr = {gf2 <- fastglm(x, y, family = binomial(), method = 1)},
fastglm.LLT = {gf3 <- fastglm(x, y, family = binomial(), method = 2)},
fastglm.LDLT = {gf4 <- fastglm(x, y, family = binomial(), method = 3)},
fastglm.qr.fpiv = {gf5 <- fastglm(x, y, family = binomial(), method = 4)},
times = 25L
)
autoplot(ct, log = FALSE) + stat_summary(fun.y = median, geom = 'point', size = 2)
# comparison of estimates
c(glm_vs_fastglm_qrcpiv = max(abs(coef(gl1) - gf1$coef)),
glm_vs_fastglm_qr = max(abs(coef(gl1) - gf2$coef)),
glm_vs_fastglm_qrfpiv = max(abs(coef(gl1) - gf5$coef)),
glm_vs_fastglm_LLT = max(abs(coef(gl1) - gf3$coef)),
glm_vs_fastglm_LDLT = max(abs(coef(gl1) - gf4$coef)))
## glm_vs_fastglm_qrcpiv glm_vs_fastglm_qr glm_vs_fastglm_qrfpiv
## 2.590289e-14 2.546921e-14 2.776945e-14
## glm_vs_fastglm_LLT glm_vs_fastglm_LDLT
## 1.140078e-13 1.094264e-13
# now between glm and speedglm
c(glm_vs_speedglm_eigen = max(abs(coef(gl1) - sg1$coef)),
glm_vs_speedglm_Chol = max(abs(coef(gl1) - sg2$coef)),
glm_vs_speedglm_qr = max(abs(coef(gl1) - sg3$coef)))
## glm_vs_speedglm_eigen glm_vs_speedglm_Chol glm_vs_speedglm_qr
## 1.359413e-12 1.359413e-12 1.191977e-12
Stability
The fastglm package does not compromise computational stability for speed. In fact, for many situations where glm() and even glm2() do not converge, fastglm() does converge.
As an example, consider the following data scenario, where the response distribution is (mildly) misspecified, but the link function is quite badly misspecified. In such scenarios, the standard IRLS algorithm tends to have convergence issues. The glm2() package was designed to handle such cases, however, it still can have convergence issues. The fastglm() package uses a similar step-halving technique as glm2(), but it starts at better initialized values and thus tends to have better convergence properties in practice.
set.seed(1)
x <- matrix(rnorm(10000 * 100), ncol = 100)
y <- (exp(0.25 * x[,1] - 0.25 * x[,3] + 0.5 * x[,4] - 0.5 * x[,5] + rnorm(10000)) ) + 0.1
system.time(gfit1 <- fastglm(cbind(1, x), y, family = Gamma(link = "sqrt")))
## user system elapsed
## 0.783 0.022 0.807
system.time(gfit2 <- glm(y~x, family = Gamma(link = "sqrt")) )
## user system elapsed
## 3.035 0.121 3.166
system.time(gfit3 <- glm2::glm2(y~x, family = Gamma(link = "sqrt")) )
## user system elapsed
## 2.117 0.084 2.205
system.time(gfit4 <- speedglm(y~x, family = Gamma(link = "sqrt")))
## user system elapsed
## 1.757 0.048 1.807
## speedglm appears to diverge
system.time(gfit5 <- speedglm(y~x, family = Gamma(link = "sqrt"), maxit = 500))
## user system elapsed
## 37.776 1.205 39.272
## Note that fastglm() returns estimates with the
## largest likelihood
c(fastglm = logLik(gfit1), glm = logLik(gfit2), glm2 = logLik(gfit3),
speedglm = logLik(gfit4), speedglm500 = logLik(gfit5))
## fastglm glm glm2 speedglm speedglm500
## -16030.81 -16704.05 -16046.66 -47722.66 -57785.72
rbind(fastglm = coef(gfit1)[1:5],
glm = coef(gfit2)[1:5],
glm2 = coef(gfit3)[1:5],
speedglm = coef(gfit4)[1:5],
speedglm500 = coef(gfit5)[1:5])
## (Intercept) X1 X2 X3 X4
## fastglm 1.429256 0.1258736 5.321164e-03 -0.1293897 0.2389373
## glm 1.431168 0.1251936 -6.896739e-05 -0.1281857 0.2366473
## glm2 1.426864 0.1242616 -9.860241e-05 -0.1254873 0.2361301
## speedglm -22.182477 3.1784570 -2.970111e+00 -4.9709797 14.0549438
## speedglm500 -27.891929 -13.9080256 -9.690833e+00 2.7279219 -11.1458325
## check convergence of fastglm and #iterations
# 1 means converged, 0 means not converged
c(gfit1$converged, gfit1$iter)
## [1] 1 17
## now check convergence for glm()
c(gfit2$converged, gfit2$iter)
## [1] 0 25
## check convergence for glm2()
c(gfit3$converged, gfit3$iter)
## [1] 1 19
## check convergence for speedglm()
c(gfit4$convergence, gfit4$iter, gfit5$convergence, gfit5$iter)
## [1] 0 25 0 500
## increasing number of IRLS iterations for glm() does not help that much
system.time(gfit2 <- glm(y~x, family = Gamma(link = "sqrt"), control = list(maxit = 1000)) )
## user system elapsed
## 116.122 4.148 120.833
gfit2$converged
## [1] FALSE
gfit2$iter
## [1] 1000
logLik(gfit1)
## 'log Lik.' -16030.81 (df=102)
logLik(gfit2)
## 'log Lik.' -16333.99 (df=102)