Induced Priors in Bayesian Mixture Models.
fipp
The goal of fipp is to provide tools to carry out sanity checks on mixture models when used for Model-Based Clustering.
Specifically, it deals with characterizing implicit quantities obtained from prior distributions of either of the following three models: Dirichlet Process Mixtures (DPM), Static Mixture of Finite Mixtures (Static MFM) and Dynamic Mixture of Finite Mixtures (Dynamic MFM).
Installation
You can install the released version of fipp from CRAN with:
install.packages("fipp")
Example: the number of filled mixture components (in other words data clusters)
One of the functions in the package allows the user to obtain the number of filled mixture components. Note that it shouldn’t be confused with the number of mixture components. The former quantity is equal or less than the latter where equality holds when at least one data point is associated to any of the mixture components in the model. For details, please refer to the vignette provided in the next section.
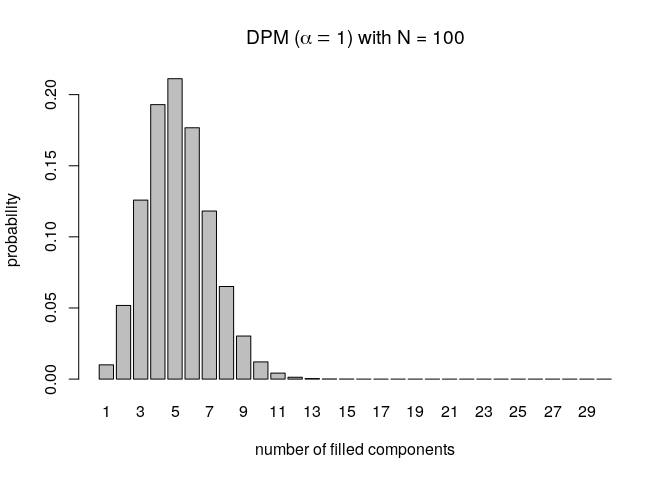
Here, we demonstrate how one can obtain the prior distribution of filled mixture components for the DPM under a specific setting (the concentration parameter (\alpha = 1) and when the sample size (N = 100)).
library(fipp)
## DPM w/ alpha = 1, N = 100, evaluate up to 30
pmfDPM <- nClusters(Kplus = 1:30, type = "DPM", alpha = 1, N = 100)
barplot(pmfDPM(),
main = expression("DPM (" * alpha == 1 * ") with N = 100"),
xlab = "number of filled components", ylab = "probability")
Details
For more detailed description regarding the functionality of the package, please refer to the vignette below: