Tools for Working with Categorical Variables (Factors).
forcats 
Overview
R uses factors to handle categorical variables, variables that have a fixed and known set of possible values. Factors are also helpful for reordering character vectors to improve display. The goal of the forcats package is to provide a suite of tools that solve common problems with factors, including changing the order of levels or the values. Some examples include:
fct_reorder(): Reordering a factor by another variable.fct_infreq(): Reordering a factor by the frequency of values.fct_relevel(): Changing the order of a factor by hand.fct_lump(): Collapsing the least/most frequent values of a factor into “other”.
You can learn more about each of these in vignette("forcats"). If you’re new to factors, the best place to start is the chapter on factors in R for Data Science.
Installation
# The easiest way to get forcats is to install the whole tidyverse:
install.packages("tidyverse")
# Alternatively, install just forcats:
install.packages("forcats")
# Or the the development version from GitHub:
# install.packages("pak")
pak::pak("tidyverse/forcats")
Cheatsheet
Getting started
forcats is part of the core tidyverse, so you can load it with library(tidyverse) or library(forcats).
library(forcats)
library(dplyr)
library(ggplot2)
starwars |>
filter(!is.na(species)) |>
count(species, sort = TRUE)
#> # A tibble: 37 × 2
#> species n
#> <chr> <int>
#> 1 Human 35
#> 2 Droid 6
#> 3 Gungan 3
#> 4 Kaminoan 2
#> 5 Mirialan 2
#> 6 Twi'lek 2
#> 7 Wookiee 2
#> 8 Zabrak 2
#> 9 Aleena 1
#> 10 Besalisk 1
#> # ℹ 27 more rows
starwars |>
filter(!is.na(species)) |>
mutate(species = fct_lump(species, n = 3)) |>
count(species)
#> # A tibble: 4 × 2
#> species n
#> <fct> <int>
#> 1 Droid 6
#> 2 Gungan 3
#> 3 Human 35
#> 4 Other 39
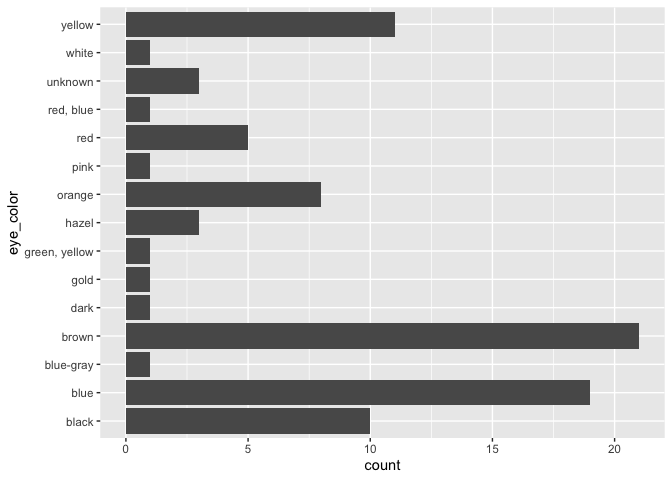
ggplot(starwars, aes(x = eye_color)) +
geom_bar() +
coord_flip()
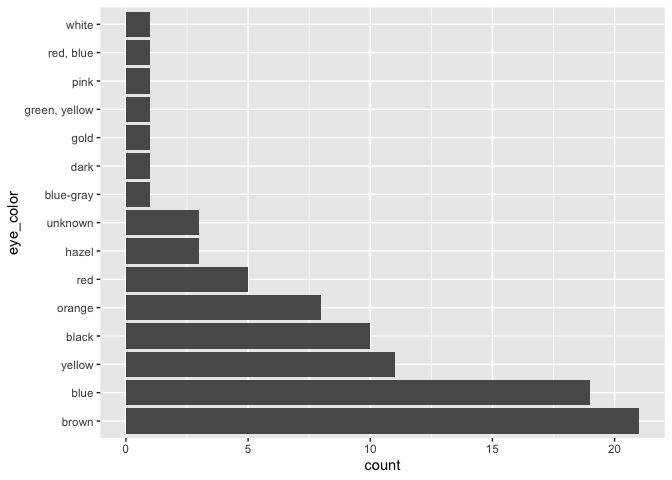
starwars |>
mutate(eye_color = fct_infreq(eye_color)) |>
ggplot(aes(x = eye_color)) +
geom_bar() +
coord_flip()
More resources
For a history of factors, I recommend stringsAsFactors: An unauthorized biography by Roger Peng and stringsAsFactors = \<sigh\> by Thomas Lumley. If you want to learn more about other approaches to working with factors and categorical data, I recommend Wrangling categorical data in R, by Amelia McNamara and Nicholas Horton.
Getting help
If you encounter a clear bug, please file a minimal reproducible example on Github. For questions and other discussion, please use https://forum.posit.co/.
