General Semiparametric Shared Frailty Model.
frailtySurv
frailtySurv is an R package for simulating and fitting semiparametric shared frailty models.
Installation
For the latest stable version, install from CRAN:
install.packages("frailtySurv")
The development version can be installed from github using devtools.
devtools::install_github("vmonaco/frailtySurv")
Example
The following code shows how to generate data and fit the model.
set.seed(1234)
library(frailtySurv)
#> Loading required package: survival
#> Welcome to frailtySurv v1.3.3
dat <- genfrail(N=200, K=2, beta=c(log(2),log(3)),
frailty="gamma", theta=2,
censor.rate=0.35,
Lambda_0=function(t, tau=4.6, C=0.01) (C*t)^tau)
fit <- fitfrail(Surv(time, status) ~ Z1 + Z2 + cluster(family),
dat=dat, frailty="gamma")
fit
#> Call: fitfrail(formula = Surv(time, status) ~ Z1 + Z2 + cluster(family),
#> dat = dat, frailty = "gamma")
#>
#> Covariate Coefficient
#> Z1 0.654
#> Z2 1.006
#>
#> Frailty distribution gamma(1.802), VAR of frailty variates = 1.802
#> Log-likelihood -1575.752
#> Converged (method) 10 iterations, 3.24 secs (maximized log-likelihood)
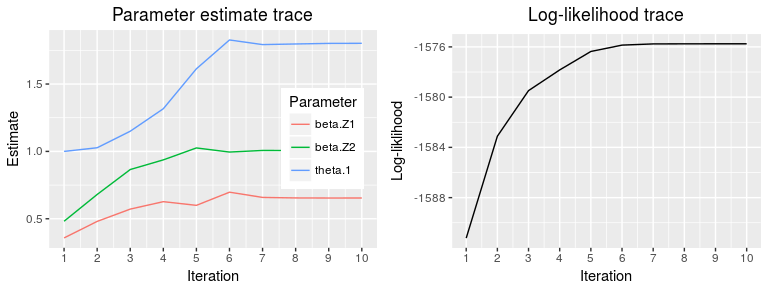
Parameter traces are given by
plot(fit, type="trace")
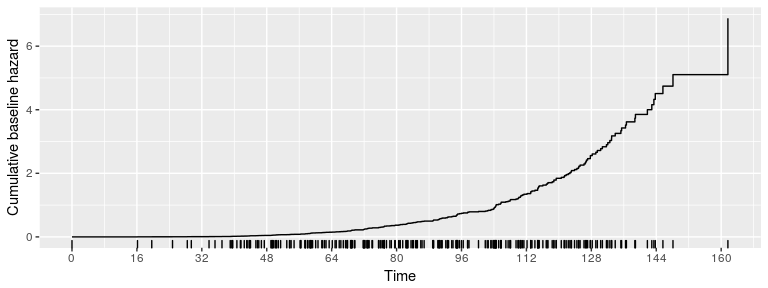
The estimated cumulative baseline hazard is given by
plot(fit, type="cumhaz")
The results can be compared to other estimation techniques.
library(survival)
library(frailtypack)
coxph(Surv(time, status) ~ Z1 + Z2 + frailty.gamma(family), data=dat)
#> Call:
#> coxph(formula = Surv(time, status) ~ Z1 + Z2 + frailty.gamma(family),
#> data = dat)
#>
#> coef se(coef) se2 Chisq DF p
#> Z1 0.6651 0.0941 0.0852 49.9190 1 1.6e-12
#> Z2 1.0282 0.1046 0.0941 96.5685 1 < 2e-16
#> frailty.gamma(family) 475.6283 150 < 2e-16
#>
#> Iterations: 7 outer, 58 Newton-Raphson
#> Variance of random effect= 1.88 I-likelihood = -1319.6
#> Degrees of freedom for terms= 0.8 0.8 149.7
#> Likelihood ratio test=576 on 151 df, p=<2e-16 n= 400
frailtyPenal(Surv(time, status) ~ Z1 + Z2 + cluster(family), data=dat, n.knots=10, kappa=2)
#>
#> Be patient. The program is computing ...
#> The program took 0.22 seconds
#> Call:
#> frailtyPenal(formula = Surv(time, status) ~ Z1 + Z2 + cluster(family),
#> data = dat, n.knots = 10, kappa = 2)
#>
#>
#> Shared Gamma Frailty model parameter estimates
#> using a Penalized Likelihood on the hazard function
#>
#> coef exp(coef) SE coef (H) SE coef (HIH) z p
#> Z1 0.67236 1.95885 0.0994732 0.0994732 6.75921 1.3875e-11
#> Z2 1.04884 2.85433 0.1191210 0.1191210 8.80481 0.0000e+00
#>
#> Frailty parameter, Theta: 1.92378 (SE (H): 0.319435 ) p = 8.5895e-10
#>
#> penalized marginal log-likelihood = -1378.15
#> Convergence criteria:
#> parameters = 0.000362 likelihood = 0.000201 gradient = 2.44e-08
#>
#> LCV = the approximate likelihood cross-validation criterion
#> in the semi parametrical case = 3.48287
#>
#> n= 400
#> n events= 256 n groups= 200
#> number of iterations: 13
#>
#> Exact number of knots used: 10
#> Value of the smoothing parameter: 2, DoF: 11.96
Clone and build
You can clone the repository and build the project from source using RStudio. To create a project in RStudio from this repository, you must have both RStudio and git installed.
- In RStudio, go to File -> New Project -> Version Control -> Git
- Name the project (eg. frailtySurv), choose location, and specify the Repository URL as
https://github.com/vmonaco/frailtySurv
See the R packages book
Build everything
To load the package (simulate installing and loading with library(frailty)):
devtools::load_all()
or shortuct: Ctrl+Shift+L
Use this command to build a source package (binary = FALSE by default)
devtools::build()
Other usefull commands and shortcuts:
- Build and reload everything: Ctrl+Shift+B
- Check the package,
devtools::check(): Ctrl+Shift+E - Run all tests,
devtools::test(): Ctrl+Shift+T
Build vignettes
Build the vignettes with the command:
devtools::build_vignettes()
Compiled vignettes will reside in inst/doc. In particular, see frailtySurv.pdf
git pull
Use RStudio to pull the latest code if you've already cloned the repository. In the "git" pane, look for the Pull button. You can also view the commit history from there and see the diff between modified files, if any.