Generalized Efficient Regression-Based Imputation with Latent Processes.
gerbil - General Efficient Regression-Based Imputation with Latent processes
This R package implements coherent multiple imputation of general multivariate data using the GERBIL algorithm described by Robbins (2020; arXiv:2008.02243).
Installation
This package is on CRAN. You are able to install the released version of gerbil from CRAN with:
install.packages("gerbil")
You can also install directly from this GitHub repository:
# run if you don't have devtools installed:
install.packages("devtools")
devtools::install_github("michaelwrobbins/gerbil")
Example
Load your dataset and run the gerbil function:
library(gerbil)
# Load the ihd sample data set with MCAR missingness:
data(ihd_mcar)
my_dataset = ihd_mcar
# Run the Imputation Process:
gerbil_object <- gerbil(dat = my_dataset, m = 1, ords = "education_level", semi = "farm_labour_days", bincat = "job_field")
#> Variable Summary:
#> Variable.Type Num.Observed Num.Miss Miss.Rate
#> sex binary 42125 0 0.00%
#> age continuous (EMP) 27382 14743 35.00%
#> marital_status binary 27382 14743 35.00%
#> job_field categorical 27382 14743 35.00%
#> farm_labour_days semicont 27382 14743 35.00%
#> own_livestock binary 27382 14743 35.00%
#> education_level ordinal 27382 14743 35.00%
#> income continuous (EMP) 27382 14743 35.00%
#>
#> Completed transformations, Time = 0.15
#> Imp. 1: gerbil initialized. Time = 2.06
#> Imp. 1: MCMC iteration 1 completed. Total time = 1.70, I-Step: 1.67, P-Step: 0.03
#> Imp. 1: MCMC iteration 2 completed. Total time = 1.81, I-Step: 1.78, P-Step: 0.03
#> Imp. 1: MCMC iteration 3 completed. Total time = 1.58, I-Step: 1.55, P-Step: 0.03
#> Imp. 1: MCMC iteration 4 completed. Total time = 1.69, I-Step: 1.67, P-Step: 0.02
#> Imp. 1: MCMC iteration 5 completed. Total time = 1.69, I-Step: 1.66, P-Step: 0.03
#> Imp. 1: MCMC iteration 6 completed. Total time = 1.75, I-Step: 1.72, P-Step: 0.03
#> Imp. 1: MCMC iteration 7 completed. Total time = 1.70, I-Step: 1.67, P-Step: 0.03
#> Imp. 1: MCMC iteration 8 completed. Total time = 1.66, I-Step: 1.63, P-Step: 0.03
#> Imp. 1: MCMC iteration 9 completed. Total time = 1.71, I-Step: 1.68, P-Step: 0.03
#> Imp. 1: MCMC iteration 10 completed. Total time = 1.53, I-Step: 1.50, P-Step: 0.03
#> Imp. 1: MCMC iteration 11 completed. Total time = 1.77, I-Step: 1.73, P-Step: 0.04
#> Imp. 1: MCMC iteration 12 completed. Total time = 1.66, I-Step: 1.63, P-Step: 0.03
#> Imp. 1: MCMC iteration 13 completed. Total time = 1.74, I-Step: 1.71, P-Step: 0.03
#> Imp. 1: MCMC iteration 14 completed. Total time = 1.78, I-Step: 1.75, P-Step: 0.03
#> Imp. 1: MCMC iteration 15 completed. Total time = 1.78, I-Step: 1.75, P-Step: 0.03
#> Imp. 1: MCMC iteration 16 completed. Total time = 1.67, I-Step: 1.64, P-Step: 0.03
#> Imp. 1: MCMC iteration 17 completed. Total time = 1.86, I-Step: 1.83, P-Step: 0.03
#> Imp. 1: MCMC iteration 18 completed. Total time = 1.61, I-Step: 1.58, P-Step: 0.03
#> Imp. 1: MCMC iteration 19 completed. Total time = 1.55, I-Step: 1.51, P-Step: 0.04
#> Imp. 1: MCMC iteration 20 completed. Total time = 1.64, I-Step: 1.61, P-Step: 0.03
#> Imp. 1: MCMC iteration 21 completed. Total time = 1.57, I-Step: 1.54, P-Step: 0.03
#> Imp. 1: MCMC iteration 22 completed. Total time = 1.58, I-Step: 1.55, P-Step: 0.03
#> Imp. 1: MCMC iteration 23 completed. Total time = 1.52, I-Step: 1.49, P-Step: 0.03
#> Imp. 1: MCMC iteration 24 completed. Total time = 1.55, I-Step: 1.53, P-Step: 0.02
#> Imp. 1: MCMC iteration 25 completed. Total time = 1.49, I-Step: 1.47, P-Step: 0.02
#> Completed untransformations for imputed dataset 1, Time = 0.04
Once you have a gerbil object, you can use the plot function to verify the quality of your imputations:
plot(gerbil_object)
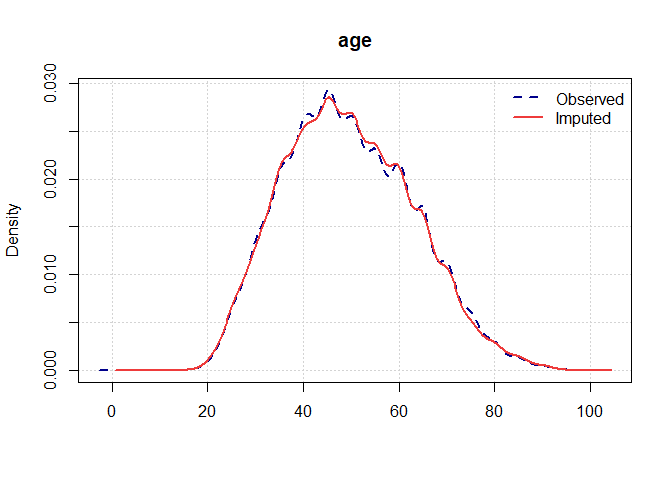
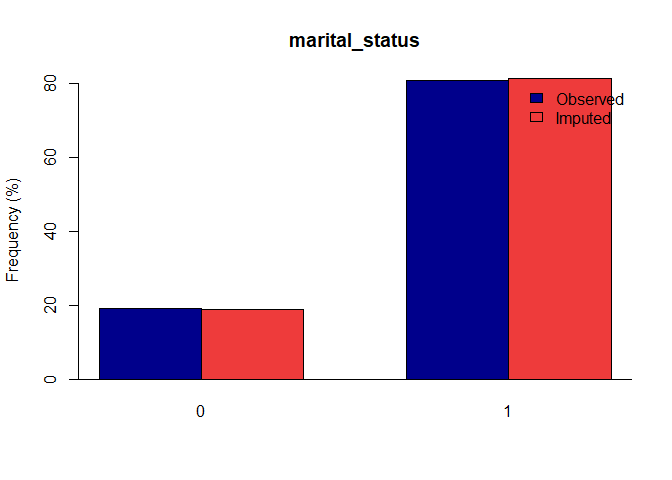
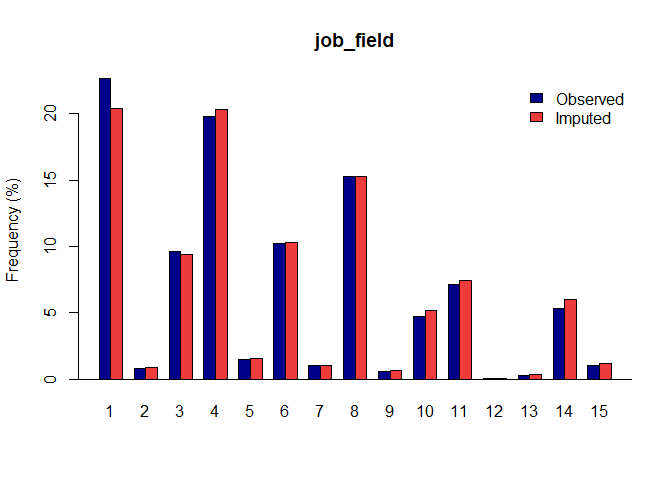
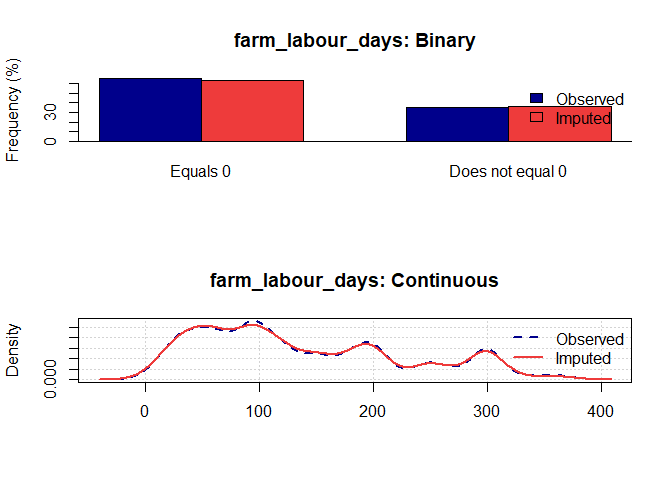
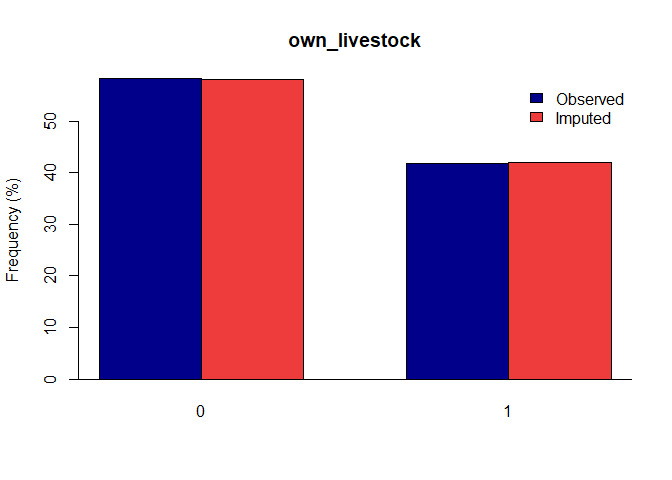
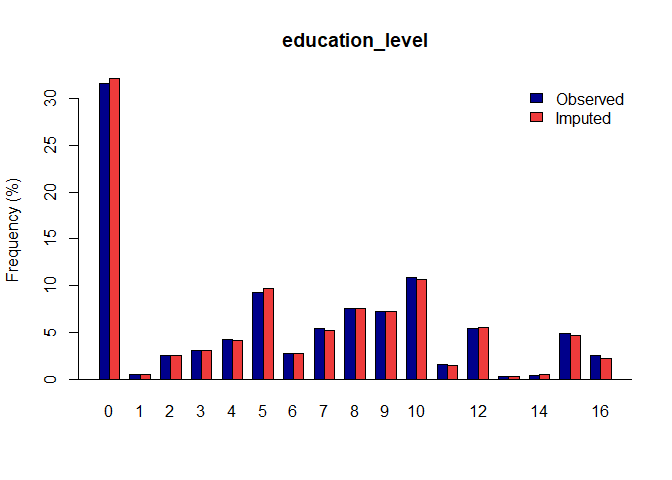
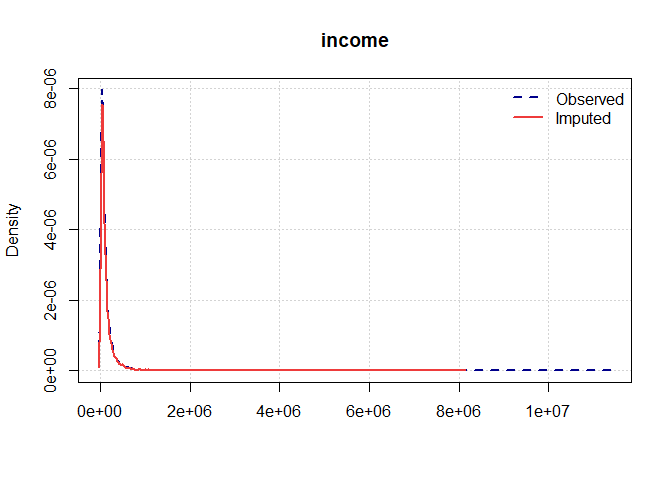
Vignettes
We have developed package vignettes that are available within the ./vignettes folder in this repository.
Tests
This package is tested at every build by the automated tests listed within the ./tests/testthat folder.
Test Coverage
One can verify our test coverage statistics by opening the r package r project and running:
# load all functions
devtools::test_coverage()

