Exploratory Symbolic Data Analysis with 'ggplot2'.
ggESDA : An R package for Exploratory Symbolic data analysis
Symbolic data analysis (SDA) is an extension of standard data analysis where symbolic data tables are used as input and symbolic objects are made output as a result. The data units are called symbolic since they are more complex than standard ones, as they not only contain values or categories, but also include internal variation and structure.[1][2]
ggESDA is an extension of ggplot2 for visualizing the symbolic data based on exploratory data analysis (EDA). The package contains many useful graphic techniques functions. Furthermore, the users can also transform the classical data into the symbolic data by the function in this package, which is one of the most important processing in SDA. For the details of the package study, you can see in Jiang and Wu(2022).
Package installation
It is recommended that download the package on CRAN, although the latest version is always upload to github first. The version in github may exist some bugs that are not fix completely.
CRAN (Update at 2022-08-17)
The ggESDA package is avaliable on CRAN. The Reference manual and Vignettes can be found there.
install.packages("ggESDA")
or
github (Update at 2022-08-17)
devtools::install_github("kiangkiangkiang/ggESDA")
After installation, the following steps will illustrate how to use the package. More details (code) about it can be found on vignettes.
Update items (only avaliable from github now)
- clustering by facets_* (index, scatter ...)
- clustering histogram using fill
- clustering indexplot using fill or group
- cov function
- sym_scale function
- fix radar problem (now it can only plot modal multi-value variables)
- update Cardiological data (correct data is Cardiological2)
- adjust text(size and color) in radar plot
- Add table output on ggInterval_hist()
- Add table output on ggInterval_2Dhist()
- change scale_sym() into scale() (generic function).
- add summary() function (generic function)
Creating
The example data is called facedata[3]. It will be the interval form with minimal and maximal, known as interval-valued data. However, most of the symbolic data are not exist at the beginning. Instead, they are usually aggregated by clustering algorithm from a classical data. Thus, we will use classic2sym() function to summarize classical data into symbolic data.
library(ggESDA)
#aggregated by the variable Species in iris
iris_interval_var <- classic2sym(iris, groupby = Species)
iris_interval_kmeans <- classic2sym(iris, groupby = "kmeans")
iris_interval_hclust <- classic2sym(iris, groupby = "hclust")
d <- data.frame(minData = runif(100, 0, 10),
maxData = runif(100, 50, 60))
d.sym <- classic2sym(d, groupby = "customize",
minData = d$minData,
maxData = d$maxData)
#get interval-valued data
d.sym$intervalData
Visualization
With the symbolic data generated, you can start to visualize the data by the following functions:
ggInterval_index() for visualizing the interval of each observations
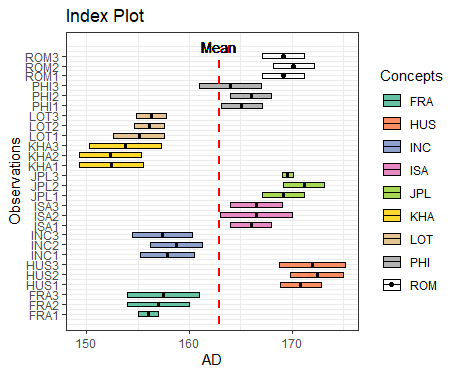
# get the mean value for the hline
m <- mean(facedata$AD)
# build a color mapping for each person
Concepts <- as.factor(rep(c("FRA", "HUS", "INC", "ISA", "JPL", "KHA",
"LOT", "PHI", "ROM"), each = 3))
# start plot
ggInterval_index(facedata, aes(x = AD, fill = Concepts))+
theme_bw() +
scale_fill_brewer(palette = "Set2")+
geom_segment(x = m, xend = m, y = 0, yend = 27,
lty = 2, col = "red", lwd = 1) +
geom_text(aes(x = m, y = 28), label = "Mean")+
scale_y_continuous(breaks = 1:27,
labels = rownames(facedata))
It can get the preliminary understanding of the interval-valued data.
You can also change fill = and col = to make the plot more visible, and set x or y axis to your variable will rotate the index line in figure.
ggInterval_minmax() for visualizing the interval of each observations which is sorted by minimal value
ggInterval_minmax(data = NULL, mapping = aes(NULL), scaleXY = "local", plotAll = F)
MMplot is an advanced graphics implemented for symbolic data, or interval-valued data. It presents the interval by the minimum and maximum, and shows the difference between each location of concept in each variable through the 45-degree line. The options scaleXY = "local" will define the axis limit for the comparsion.
ggInterval_boxplot() for visualizing the distribution of the interval data
ggInterval_boxplot(facedata, plotAll = T) + theme_bw()
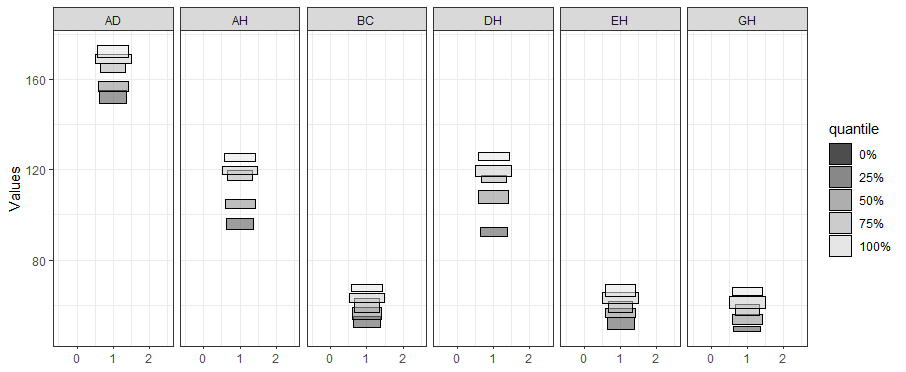
The side-by-side boxplot clearly shows the difference of variables' distribution.
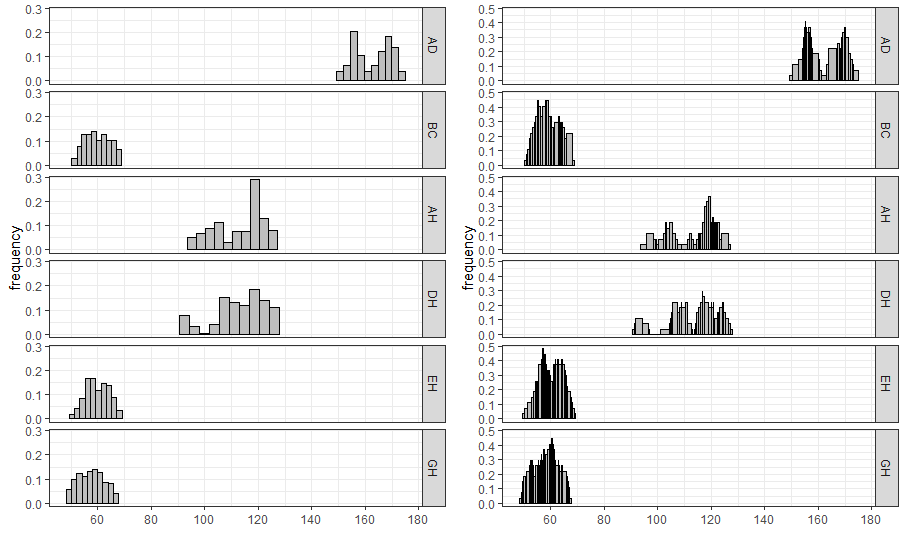
ggInterval_hist() for visualizing the distribution of the interval data
For interval-valued data, not only equidistant-bin histogram but the Non-equidistant-bin histogram will exhibit the distribution. Use the option method, and set by equal-bin or unequal-bin. See the details Jiang and Wu(2022).
equal_bin <- ggInterval_hist(facedata, plotAll = T) +
theme_bw()
unequal_bin <- ggInterval_hist(facedata, plotAll = T,
method = "unequal-bin") +
theme_bw()
ggarrange(equal_bin, unequal_bin, ncol = 2)
ggInterval_centerRange for visualizing the relation between center and range of the interval of each observations.
ggInterval_centerRange(data = NULL, mapping = aes(NULL), plotAll = F)
Another advanced graphics implemented is called center-range plot, which helps researchers to be able to grasp the relationship between center and range.
ggInterval_scatter for visualizing the the relation of two variables
A scatter plot of interval-valued data is presented by a rectangle, which is composed of two interval.
As well, ggInterval_scaMatrix is an extension for visualizing all variables relations by using scatter plot at a time.
ggInterval_scaMatrix(facedata)
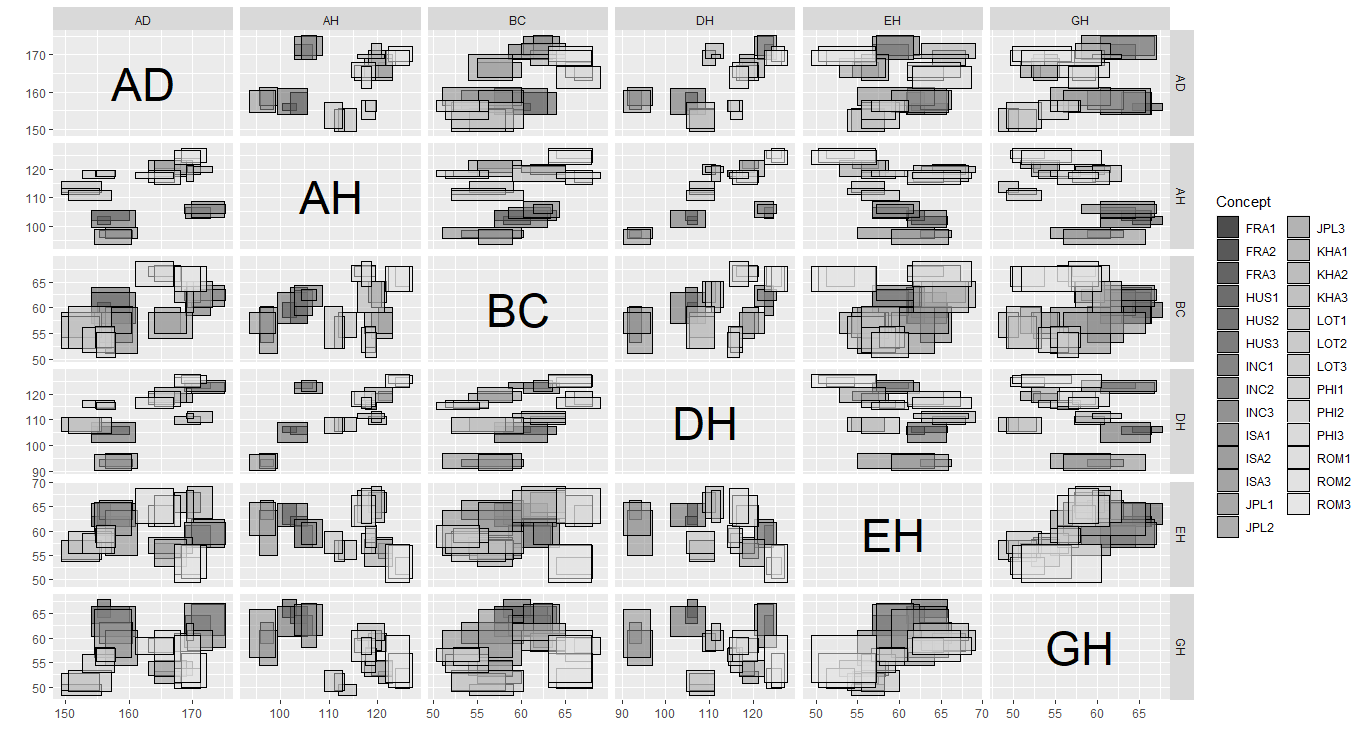
ggInterval_2Dhist for visualizing the two variable distribution by theirs frequency of interval bins
Another bivariate relationship plot is two-dimension histogram, which is to calculate joint histogram frequency and using matrix visualization to plot.
By the extension, ggInterval_2DhistMatrix is to visualize all variables relations by using 2Dhist plot at a time
ggInterval_2DhistMatrix(facedata,
xBins = 10,
yBins = 10,
removeZero = T,
addFreq = F)
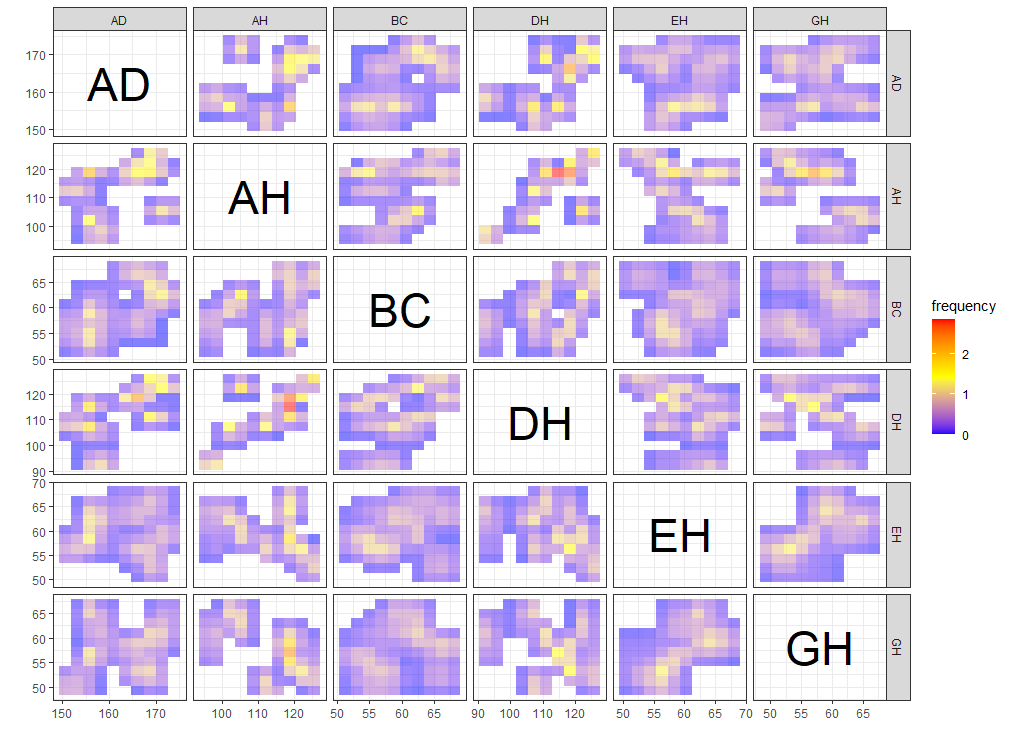
ggInterval_indexImage for visualizing the interval of each observations by using image
A heatmap type presentation for the interval by the option plotAll and full_strip. There will be two distinct type, column condition and matrix condition. See the details Jiang and Wu(2022).
p1 <- ggInterval_indexImage(facedata, plotAll = T, column_condition = T,
full_strip = T)
p2 <- ggInterval_indexImage(facedata, plotAll = T, column_condition = F,
full_strip = T)
ggpubr::ggarrange(p1, p2, ncol = 2)
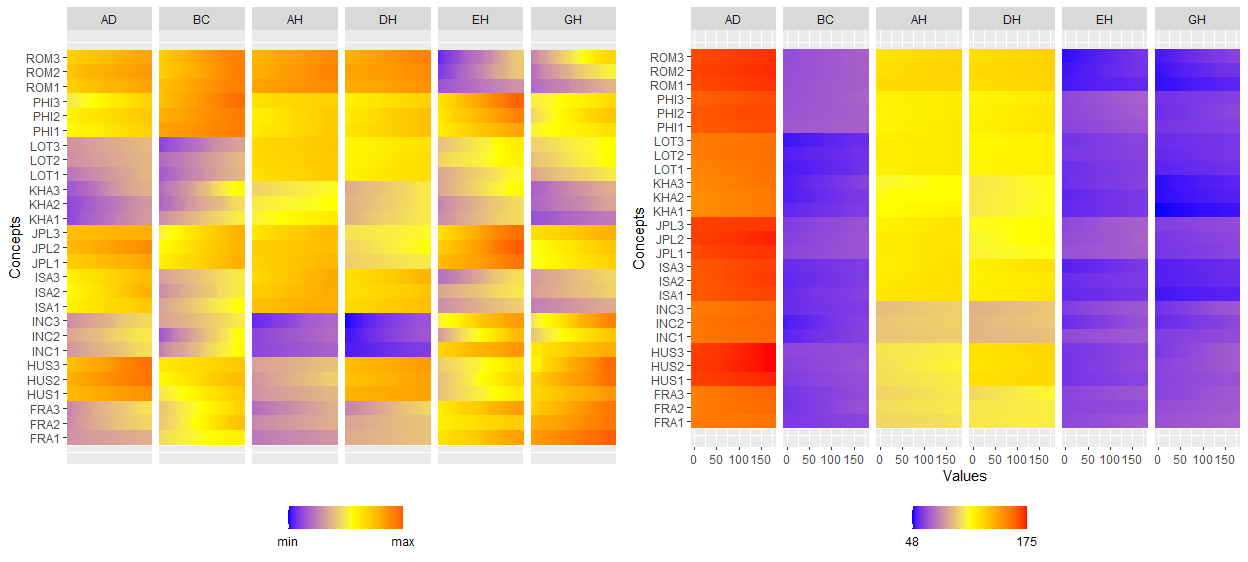
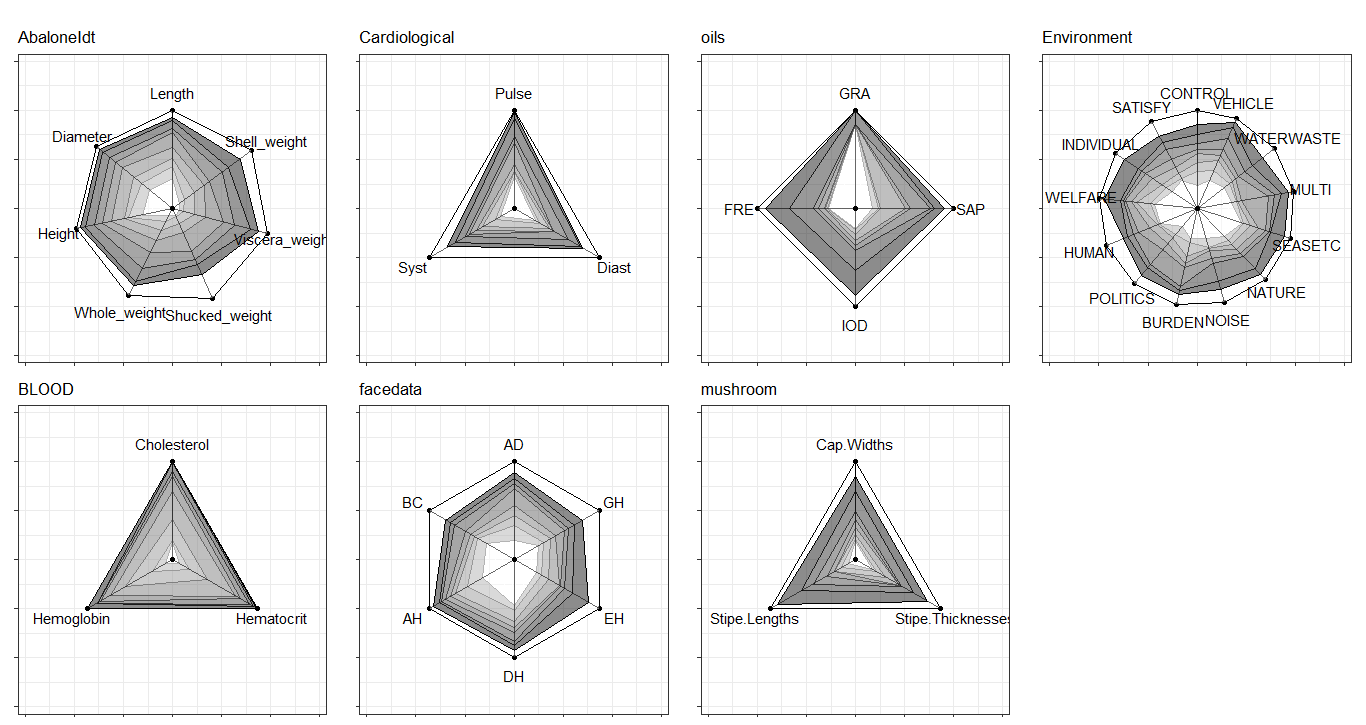
ggInterval_radar for visualizing the interval of multivariates
One of the most well-known multivariate visualization techniques is radar plot, or called start plot. We fill the interval area by color mapping, and compare CONCEPTs, surely you can arrange it.
In ggInterval_radar, you can add any annotations in figure, including a circle for classify the normalize data position, a real value for the interval or a propotion for modal multi-valued variables. There is the most important options plotPartial, which define which observations you want to plot. For example, ggInterval_radar(data = facedata, plotPartial = c(1, 3)). It means we want to plot the observations whose position of row is at 1 and 3. Remark: A radar plot will visualize all variables you on your input data in one time. If you only want to show the partial variables, just clarify in your input data, ex data = facedata[, 1:3]. However, the data you input must have full observations in order to normalize, so an obvious ERROR for input options in data is a data = facedata[1:5, ], which only contains the first five data not a full. Unless you want to make them be your population.
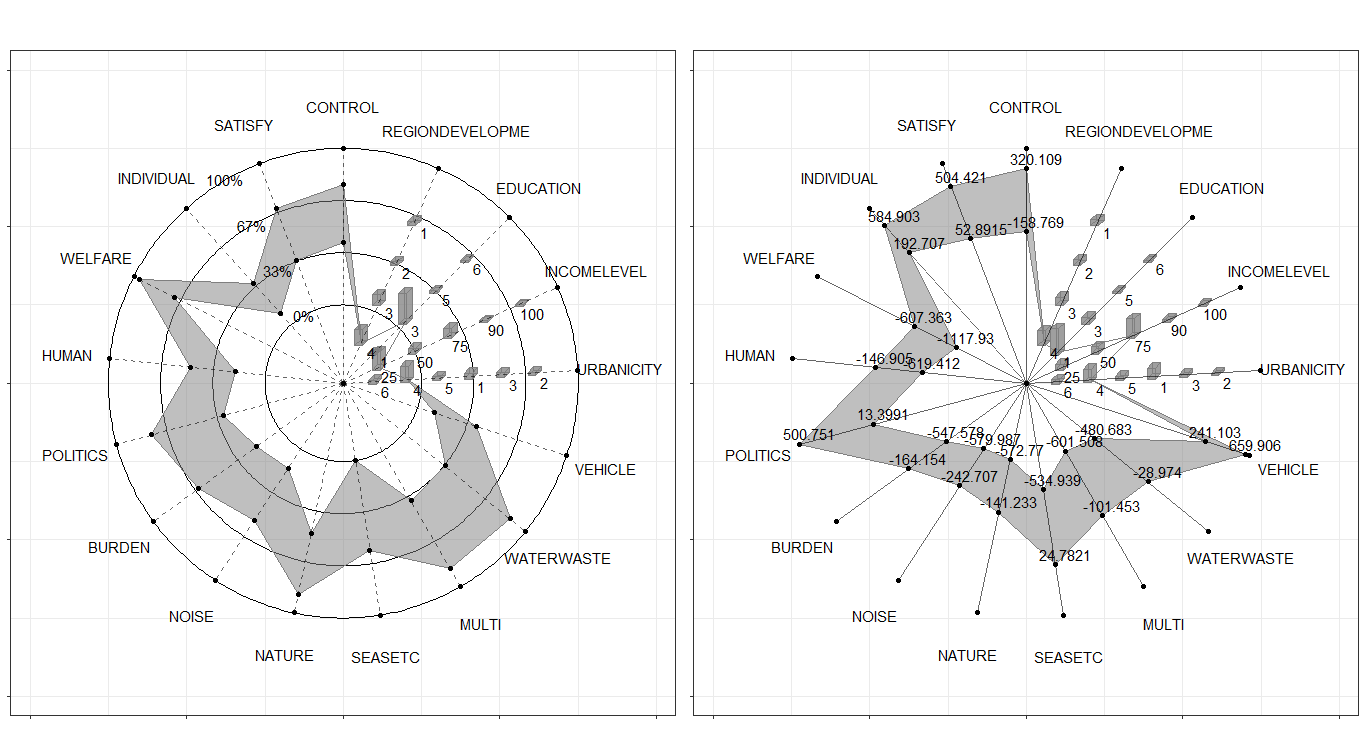
Surely, we always compare the data in the same figure. Just use the color mapping for the different observations.
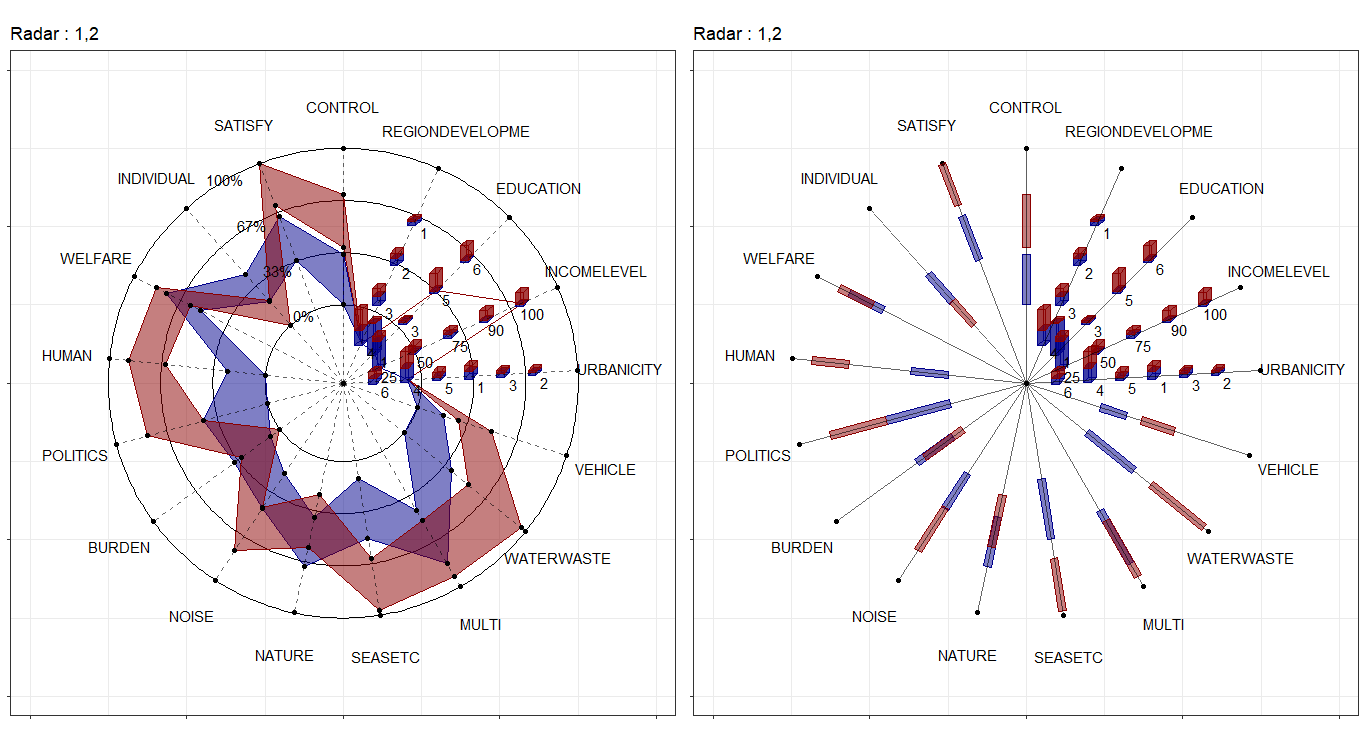
In the field of interval-valued data, the quantile is usually for analysis. We can present the quantiles for each datasets.
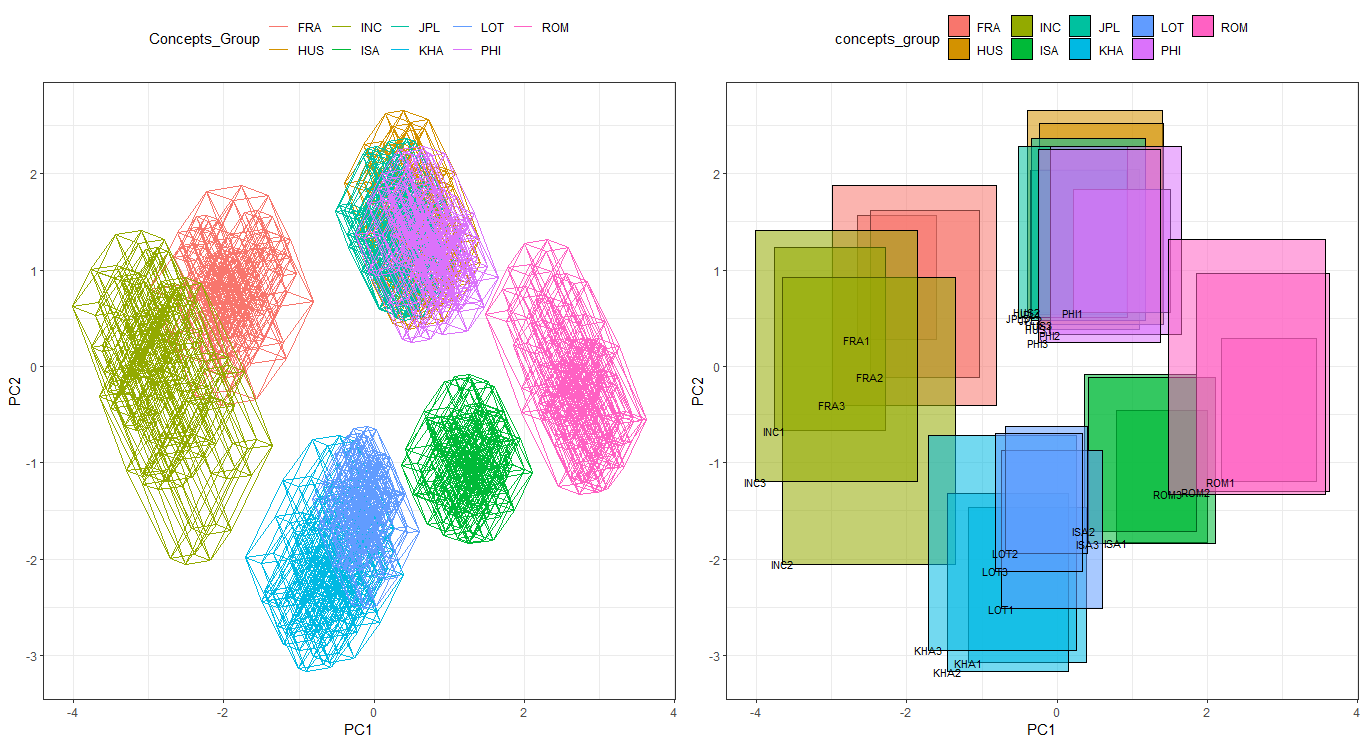
ggInterval_PCA for dimension reduction in interval data
Two kinds of dimension reduction, see the details Jiang and Wu(2022), show in following:
CONCEPT <- rep(c("FRA", "HUS", "INC", "ISA", "JPL", "KHA",
"LOT", "PHI", "ROM"), each = 3)
p <- ggInterval_PCA(facedata, poly = T,
concepts_group = CONCEPT)
p$ggplotPCA <- p$ggplotPCA + theme(legend.position = "top") +
theme_bw()
p2 <- ggInterval_PCA(facedata, poly = F,
concepts_group = CONCEPT)
p2$ggplotPCA <- p2$ggplotPCA + theme(legend.position = "top") +
theme_bw()
ggpubr::ggarrange(p$ggplotPCA, p2$ggplotPCA, ncol = 2)
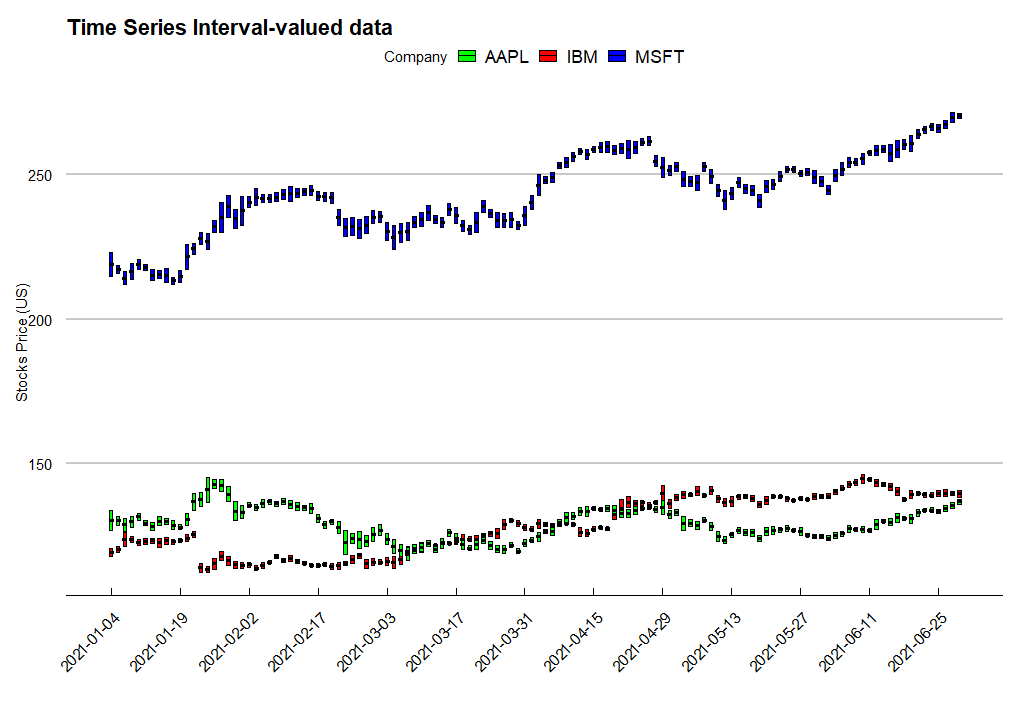
Time series interval-valued data
In practice, we can use the technique to visualize financial data, such as stocks data. We get the stocks price of AAPL, MSFT and IBM from 2021-01-04 to 2021-12-31. Use the function ggInterval_index, and we can easily present the daily low and high price.
Note: you must specify the classification variable, whose length is equal to the data you input, in fill when you want to classify. The classification variable may not be in your input data frame as the following code (Company variable is out of the input data d.i). Each length of the classification factor must be equal as well.
library(data.table)
library(ggthemes)
# Read data (from 2021-01-04 to 2021-12-31)
ibm <- fread("https://query1.finance.yahoo.com/v7/finance/download/IBM?period1=1609459200&period2=1640995200&interval=1d&events=history&includeAdjustedClose=true")
msft <- fread("https://query1.finance.yahoo.com/v7/finance/download/MSFT?period1=1609459200&period2=1640995200&interval=1d&events=history&includeAdjustedClose=true")
aapl <- fread("https://query1.finance.yahoo.com/v7/finance/download/AAPL?period1=1609459200&period2=1640995200&interval=1d&events=history&includeAdjustedClose=true")
# Get half year (1~6 months) for demo : 124 records
d1 <- data.frame(min = ibm[1:124, "Low"],
max = ibm[1:124, "High"])
d2 <- data.frame(min = msft[1:124, "Low"],
max = msft[1:124, "High"])
d3 <- data.frame(min = aapl[1:124, "Low"],
max = aapl[1:124, "High"])
# Combine three company and transform to symbolic data
d <- rbind(d1, d2, d3)
d.i <- classic2sym(d, groupby = "customize",
minData = d$min,
maxData = d$max)$intervalData
# Make classification variable
Company <- rep(c("IBM", "MSFT", "AAPL"), each = 124)
# plot full data and classify by Company (fill)
ggInterval_index(d.i, aes(y = V1, fill = Company, group = Company)) +
scale_x_continuous(breaks = seq(1, 124, 10),
labels = c(ibm[seq(1, 124, 10), 1])$Date)+
labs(x = "", y = "Stocks Price (US)", title = "Time Series Interval-valued data") +
scale_fill_manual(values = c("green", "red", "blue")) +
theme_economist_white(gray_bg = FALSE) +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))
References
1. Diday, Edwin; Esposito, Floriana (December 2003). "An introduction to symbolic data analysis and the SODAS software".
2. Lynne Billard; Edwin Diday (14 May 2012). Symbolic Data Analysis: Conceptual Statistics and Data Mining.
3. Billard L. and Diday E. (2006). Symbolic data analysis: Conceptual statistics and data mining. Wiley, Chichester.