Graphs for Correspondence Analysis.
ggfacto
Readable, complete and pretty graphs for correspondence analysis made with FactoMineR. Many can be rendered as interactive html plots, showing useful informations at mouse hover. The interest is not mainly visual but statistical : it helps the reader to keep in mind the data contained in the cross-table or Burt table while reading correspondence analysis, thus preventing overinterpretation. Graphs are made with ggplot2, which means that you can use the + syntax to manually add as many graphical pieces you want, or change theme elements.
Installation
You can install ggfacto from CRAN:
install.packages("ggfacto")
Or install the development version from github:
# install.packages("devtools")
devtools::install_github("BriceNocenti/ggfacto")
Interactive plot from multiple correspondence analysis (MCA)
Make the MCA (using a wrapper function around FactoMineR:MCA) :
library(ggfacto)
data(tea, package = "FactoMineR")
res.mca <- MCA2(tea, active_vars = 1:18)
Make the plot (as a ggplot2 object) and add a supplementary variable ("SPC") :
graph_mca <- ggmca(res.mca, tea, sup_vars = "SPC", profiles = TRUE, text_repel = TRUE)
Use text_repel = TRUE to avoid overlapping of text, and obtain a more readable image (be careful that, if the plot is overloaded, labels can be far away from their original location).
Use profiles = TRUE to draw the graph of individuals : one point is added for each profile of answers.
Turn the plot interactive :
ggi(graph_mca)
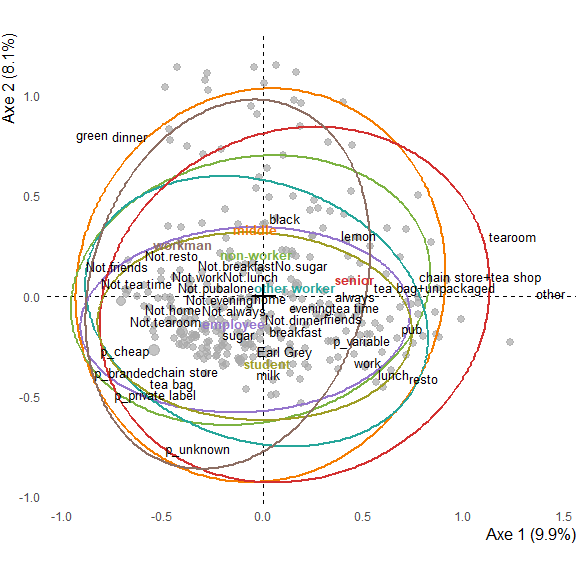
See the crosstables of active variables directly on the plot
It is possible to print all crosstables between active variables (burt table) into the interactive tooltips. Spread from mean are colored and, usually, points near the middle will have less colors, and points at the edges will have plenty. It may takes time to print, but really helps to interpret the MCA in close proximity with the underlying data.
ggmca(res.mca, tea, sup_vars = "SPC", active_tables = "active",
ylim = c(NA, 1.2), text_repel = TRUE) %>%
ggi()
See the distribution of active variables for each level of a supplementary variable
ggmca(res.mca, tea, sup_vars = "SPC", active_tables = "sup",
ylim = c(NA, 1.2), text_repel = TRUE) %>%
ggi()
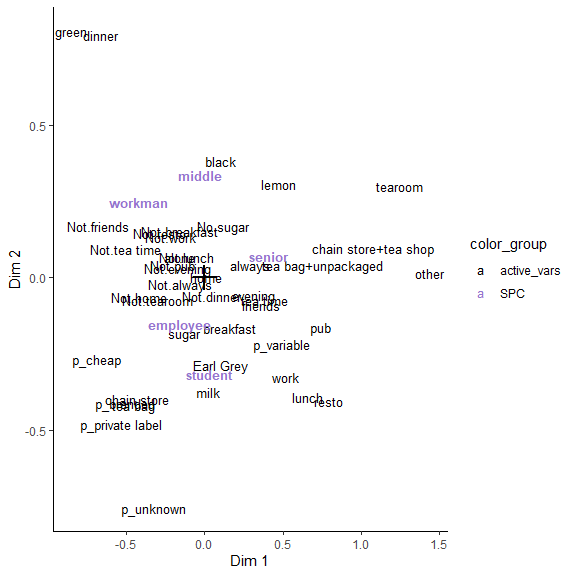
Concentration ellipses for each levels of a supplementary variable
ggmca(res.mca, tea, sup_vars = "SPC", ylim = c(NA, 1.2), ellipses = 0.95, text_repel = TRUE, profiles = TRUE)
#> colors based on the following categories (rename with colornames_recode): 'SPC_employee', 'SPC_middle', 'SPC_non-worker', 'SPC_other worker', 'SPC_senior', 'SPC_student', 'SPC_workman'
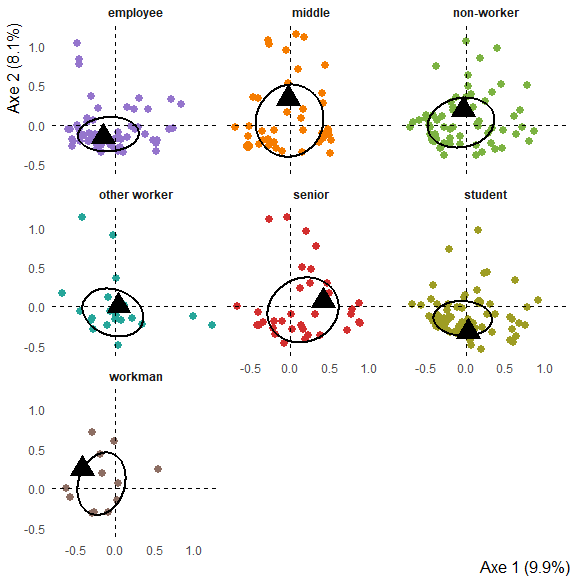
Graph of profiles of answer for each levels of a supplementary variable (with median ellipses containing half the population)
ggmca(res.mca, tea, sup_vars = "SPC", ylim = c(NA, 1.2), type = "facets", ellipses = 0.5, profiles = TRUE)
#> colors based on the following categories (rename with colornames_recode): 'SPC_employee', 'SPC_middle', 'SPC_non-worker', 'SPC_other worker', 'SPC_senior', 'SPC_student', 'SPC_workman'
Simple correspondence analysis (CA)
Make the correspondence analysis :
tabs <- tabxplor::tab_plain(forcats::gss_cat, race, marital, df = TRUE)
res.ca <- FactoMineR::CA(tabs, graph = FALSE)
Interactive plot :
graph.ca <- ggca(res.ca,
title = "Race by marical : correspondence analysis",
tooltips = c("row", "col"))
ggi(graph.ca)
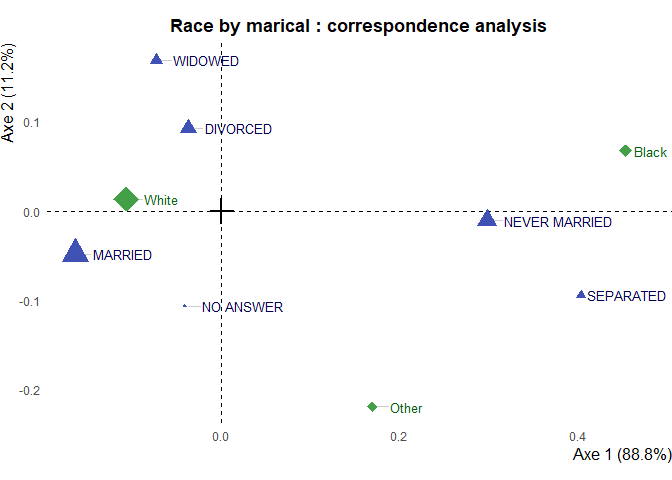
Image plot (with text_repel to avoid overlapping of labels) :
ggca(res.ca,
title = "Race by marical : correspondence analysis",
text_repel = TRUE, dist_labels = 0.02)
Personnalize plots
Step-by-step functions can be used to create a database with all the necessary data, modify it, then use it to draw the plot:
library(dplyr)
library(ggplot2)
plot_data <- ggmca_data(res.mca, tea, sup_vars = "SPC")
plot_data$vars_data <- plot_data$vars_data %>%
filter(!lvs %in% c("other worker", "non-worker"))
ggmca_plot(plot_data, ylim = c(NA, 1.2), text_repel = TRUE)
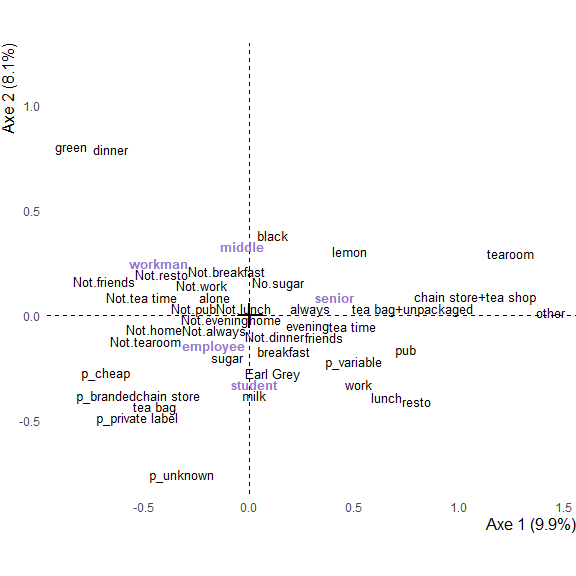
The plot can always be modified using the ggplot2+ operator :
ggmca_plot(plot_data, ylim = c(NA, 1.2)) +
labs(title = "Multiple correspondence analysis") +
theme(axis.line = element_line(linetype = "solid") )
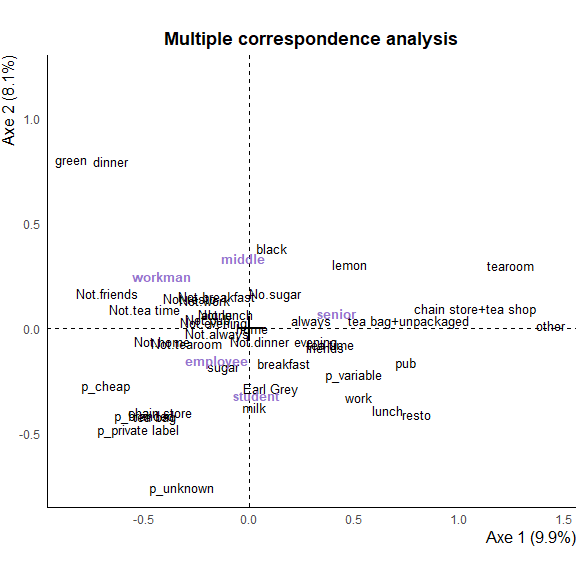 You can then pass to plot to
You can then pass to plot to ggi() to make it interactive.
Set use_theme = FALSE to use you own ggplot2 theme :
ggmca_plot(plot_data, ylim = c(NA, 1.2), use_theme = FALSE) +
theme_classic()