Description
Dots and Their Connections in Polar Coordinate System.
Description
Provides basic graphing functions to fully demonstrate point-to-point connections in a polar coordinate space.
README.md
ggpolar: Dots and Their Connections in Polar Coordinate System
{ggpolar} provides a very flexible way to create dots in coordinate system for event list and connect the dots with segments based on {ggplot2}.
Installation
You can install the released version of {ggpolar} from CRAN with:
install.packages("ggpolar")
You can install the development version of {ggpolar} from GitHub with:
remotes::install_github("ShixiangWang/polar")
Example
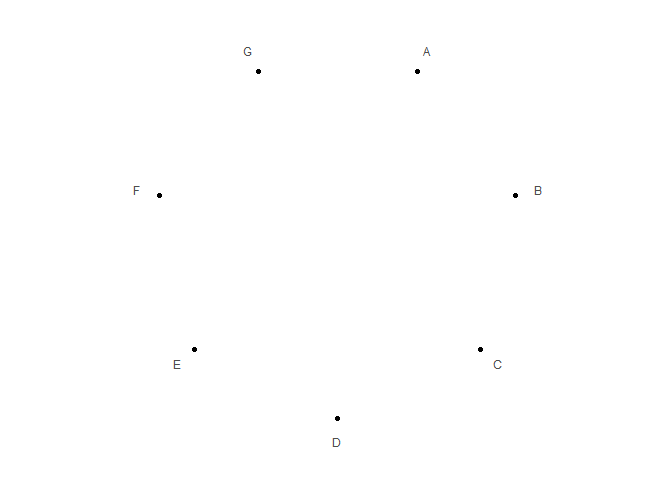
Init a polar plot
library(ggpolar)
#> Loading required package: ggplot2
data <- data.frame(x = LETTERS[1:7])
p1 <- polar_init(data, x = x)
p1
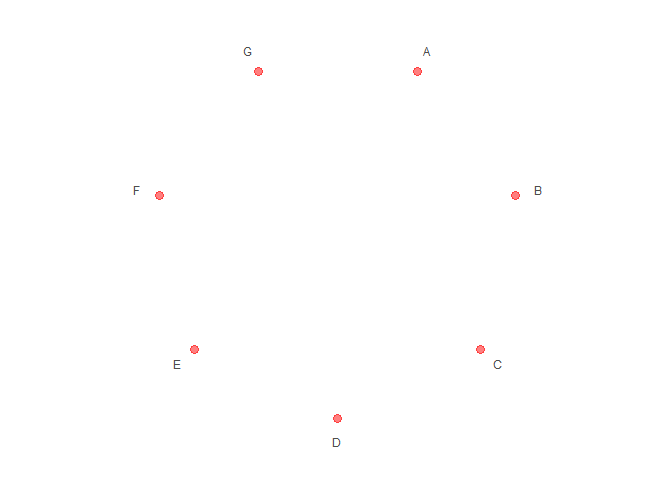
# Set aes value
p2 <- polar_init(data, x = x, size = 3, color = "red", alpha = 0.5)
p2
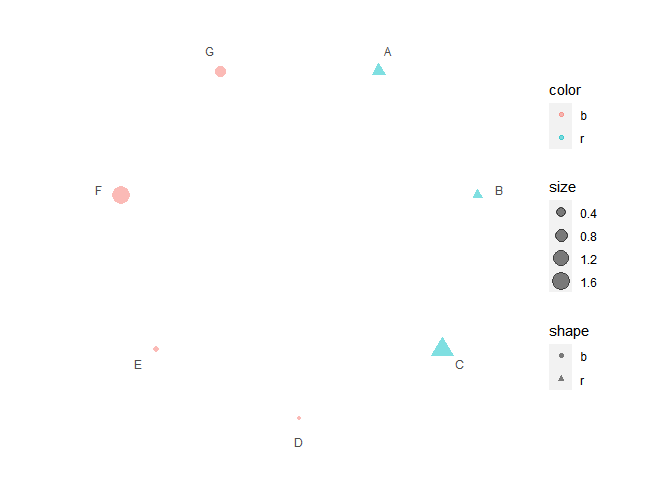
# Set aes mapping
set.seed(123L)
data1 <- data.frame(
x = LETTERS[1:7],
shape = c("r", "r", "r", "b", "b", "b", "b"),
color = c("r", "r", "r", "b", "b", "b", "b"),
size = abs(rnorm(7))
)
# Check https://ggplot2.tidyverse.org/reference/geom_point.html
# for how to use both stroke and color
p3 <- polar_init(data1, x = x, aes(size = size, color = color, shape = shape), alpha = 0.5)
p3
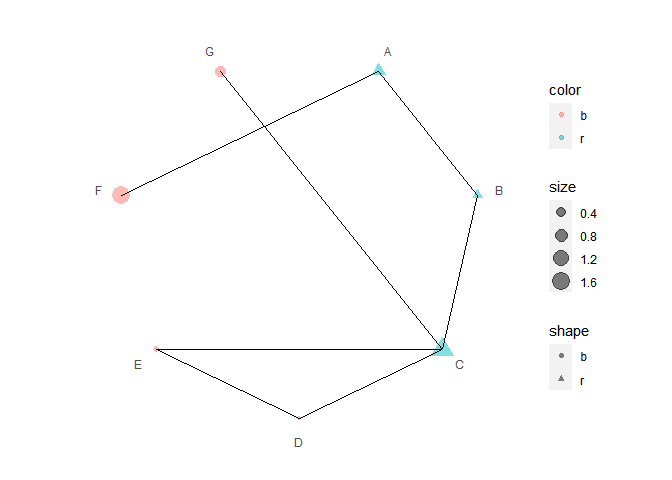
Connect polar dots
data2 <- data.frame(
x1 = LETTERS[1:7],
x2 = c("B", "C", "D", "E", "C", "A", "C"),
color = c("r", "r", "r", "b", "b", "b", "b")
)
p4 <- p3 + polar_connect(data2, x1, x2)
p4
# Unlike polar_init, mappings don't need to be included in aes()
p5 <- p3 + polar_connect(data2, x1, x2, color = color, alpha = 0.8, linetype = 2)
p5
# Use two different color scales
if (requireNamespace("ggnewscale")) {
library(ggnewscale)
p6 = p3 +
new_scale("color") +
polar_connect(data2, x1, x2, color = color, alpha = 0.8, linetype = 2)
print(p6 + scale_color_brewer())
print(p6 + scale_color_manual(values = c("darkgreen", "magenta")))
}
#> Loading required namespace: ggnewscale
#> Warning: package 'ggnewscale' was built under R version 4.2.1
Citation
If you use {ggpolar} in academic research, please cite the following paper along with the GitHub repo.
Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction, eLife. https://doi.org/10.7554/eLife.49020.