Grammar Extensions to 'ggplot2'.
ggpp 
Purpose
Package ‘ggpp’ provides a set of building blocks that extend the Grammar of Graphics implemented in package ‘ggplot2’ (>= 3.5.0). The extensions enhance the support of data labels and annotations in plots. Position functions implement new approaches to nudging usable with any geometry, but especially useful together with geom_text_s() and geom_label_s() from this package and geom_text_repel() and geom_label_repel() from package ‘ggrepel’ (>= 0.9.2). See: (https://ggrepel.slowkow.com) for installation instructions and news about releases.
NPC support in ggplot2
Normalised Parent Coordinates (NPC) are supported natively by ‘ggplot2’ >= 3.5.0 by means of R’s identity function I(). This new approach does not require special geometries as it should work with almost every existing geometry. However, currently this works only when continuous variables are mapped to the x and/or y aesthetics. If this mechanism is extended to support factors and other discrete variables in the future , all the _npc geometries from ‘ggpp’ could become redundant. Meanwhile, these special geometries from ‘ggpp’ still fill a gap, albeit a smaller one, in the support of NPC by ‘ggplot2’.
Extended Grammar of graphics
Geometries
The distinction between observations or data mapped to x and y aesthetics and data labels is that data labels are linked to a the coordinates of the data, but their own location is usually nearby but not exactly that of the data. In other words the location of a data label in x and y coordinates is flexible as long as the link to a data observation can be inferred. In the case of annotations the location on the plotting area is arbitrary, dictated by available graphic design considerations and the requirement of not occluding data observations. In the table below we list for the geometries defined in package ‘ggpp’: 1) whether they are intended for data labels, annotations or data, 2) the aesthetics and pseudo-aesthetics they obey, and 3) whether they can connect the original data position to the displaced position. The drawing of connecting segments or arrows between the displaced and original positions, those of the observation and the displaced label, requires also a change in the data returned by position functions (see the next section).
| Geometry | Main use | Aesthetics | Segment |
|---|---|---|---|
geom_text_s() | data labels | x, y, label, size, family, font face, colour, alpha, group, angle, vjust, hjust | yes |
geom_label_s() | data labels | x, y, label, size, family, font face, colour, fill, alpha, linewidth, linetype, group, vjust, hjust | yes |
geom_text_pairwise() | data labels | x, xmin, xmax, y, label, size, family, font face, colour, alpha, group, angle, vjust, hjust | horiz. |
geom_label_pairwise() | data labels | x, xmin, xmax, y, label, size, family, font face, colour, fill, alpha, linewidth, linetype, group, vjust, hjust | horiz. |
geom_point_s() | data labels | x, y, size, colour, fill, alpha, shape, stroke, group | yes |
geom_table()1 | data labels | x, y, label, size, family, font face, colour, alpha, group, angle, vjust, hjust | yes |
geom_plot()1, geom_grob()1 | data labels | x, y, label, group, angle, vjust, hjust | yes |
geom_margin_arrow() | data labels, scale labels, data | xintercept, yintercept, label, size, family, font face, colour, alpha, group, vjust, hjust | no |
geom_margin_point() | data labels, scale labels, data | xintercept, yintercept, label, size, family, font face, colour, alpha, group, vjust, hjust | no |
geom_margin_grob() | data labels, scale labels, data | xintercept, yintercept, label, size, family, font face, colour, alpha, group, vjust, hjust | no |
geom_quadrant_lines() , geom_vhlines() | data labels, scale labels, data | xintercept, yintercept, label, size, family, font face, colour, alpha, group, vjust, hjust | no |
Geometries defined in package ‘ggpp’. 1 NPC versions exist for these geometries, as well as for geom_text() and geom_label(), used mainly for plot annotations.
Position functions
In contrast to position functions from ‘ggplot2’ all the position functions from package ‘ggpp’ are able keep the original x and y coordinates under a different name in the data object when displacing them to a new position. This makes them compatible with geom_text_s(), geom_label_s(), geom_point_s(), geom_table(), geom_plot() and geom_grob() from this package. All these geoms can draw segments or arrows connecting the original positions to the displaced positions. They remain backwards compatible and can be used in all geometries that have a position formal parameter. This is similar to the approach used in package ‘ggrepel’ (<= 0.9.1) but uses a different naming convention that allows the new position functions to remain backwards compatible with ‘ggplot2’. Starting from version 0.9.2 geometries geom_text_repel() and geom_label_repel() from package ‘ggrepel’ are fully compatible with this new naming convention.
Position functions position_nudge_keep(), position_nudge_to(), position_nudge_center() and position_nudge_line() implement different flavours of nudging. The last two functions make it possible to apply nudging that varies automatically according to the relative position of data points with respect to arbitrary points or lines, or with respect to a polynomial or smoothing spline fitted on-the-fly to the the observations.
Position functions position_stacknudge(), position_fillnudge(), position_jitternudge(), position_dodgenudge() and position_dodge2nudge() combine each the roles of two position functions. They make it possible to easily nudge labels in plot layers that use stacking, dodging or jitter. Functions position_jitter_keep(), position_stack_keep(), position_fill_keep(), position_dodge_keep(), position_dosge2_keep() behave like the positions from ‘ggplot2’ but keep in the data object the original coordinates.
| Position | Main use | Displacement | Most used with |
|---|---|---|---|
position_nudge_keep() | nudge | x, y (fixed distance) | data labels |
position_jitter_keep() | jitter | x, y (random) | dot plots |
position_stack_keep() | stack | vertical (absolute) | column and bar plots |
position_stack_minmax() | stack | vertical (absolute) | error bars |
position_fill_keep() | fill | vertical (relative, fractional) | column plots |
position_dodge_keep() | dodge | sideways (absolute) | column and bar plots |
position_dosge2_keep() | dodge2 | sideways (absolute) | box plots |
position_nudge_to() | nudge | x, y (fixed position) | data labels |
position_nudge_center() | nudge | x, y (away or towards target) | data labels |
position_nudge_line() | nudge | x, y (away or towards target) | data labels |
position_stacknudge() | stack + nudge | combined, see above | data labels in column plots |
position_fillnudge() | fill + nudge | combined, see above | data labels in column plots |
position_jitternudge() | jitter + nudge | combined, see above | data labels in dot plots |
position_dodgenudge() | dodge + nudge | combined, see above | data labels in column plots |
position_dodge2nudge() | dodge2 + nudge | combined, see above | data labels in box plots |
Position functions defined in package ‘ggpp’.
Statistics
Statistic stat_fmt_tb() helps with the formatting of tables to be plotted with geom_table().
Four statistics, stat_dens2d_filter(), stat_dens2d_label(), stat_dens1d_filter() and stat_dens1d_label(), implement tagging or selective labeling of observations based on the local 2D density of observations in a panel. Another two statistics, stat_dens1d_filter_g() and stat_dens1d_filter_g() compute the density by group instead of by plot panel. These six statistics are designed to work well together with geom_text_repel() and geom_label_repel() from package ‘ggrepel’ (>= 0.8.0).
The statistics stat_apply_panel() and stat_apply_group() are useful for applying arbitrary functions returning numeric vectors like cumsum(), cummax() and diff(). Statistics stat_centroid() and stat_summary_xy() allow computation of summaries on both x and y and passing them to a geom.
The statistics stat_quadrant_counts() and stat_panel_counts() make it easy to annotate plots with the number of observations.
| Statistic | Main use | Usual geometries | Most used with | Compute function |
|---|---|---|---|---|
stat_fmt_tb() | formatting and selection | geom_table() | tables as data labels | group |
stat_fmt_tb() | formatting and selection | geom_table_npc() | tables as annotations | group |
stat_dens2d_filter() | local 2D density filtering | geom_text_s(), geom_label_s(), geom_text(), geom_label() | text as data labels | panel |
stat_dens2d_label() | local 2D density filtering | geom_text_repel(), geom_label_repel() | text as data labels | panel |
stat_dens1d_filter() | local 1D density filtering | geom_text_s(), geom_label_s(), geom_text(), geom_label() | text as data labels | panel |
stat_dens1d_label() | local 1D density filtering | geom_text_repel(), geom_label_repel() | text as data labels | panel |
stat_dens2d_filter_g() | local 2D density filtering | geom_text_s(), geom_label_s(), geom_text(), geom_label() | text as data labels | group |
stat_dens2d_label_g() | local 2D density filtering | geom_text_repel(), geom_label_repel() | text as data labels | group |
stat_dens1d_filter_g() | local 1D density filtering | geom_text_s(), geom_label_s(), geom_text(), geom_label() | text as data labels | group |
stat_dens1d_label_g() | local 1D density filtering | geom_text_repel(), geom_label_repel() | data labels | group |
stat_panel_counts() | number of observations | geom_text(), geom_label() | text as annotation | panel |
stat_group_counts() | number of observations | geom_text(), geom_label() | text as annotation | panel |
stat_quadrant_counts() | number of observations | geom_text(), geom_label() | text as annotation | panel |
stat_apply_panel() | cummulative summaries | geom_point(), geom_line(), etc. | scatter and line plots | panel |
stat_apply_group() | cummulative summaries | geom_point(), geom_line(), etc. | scatter and line plots | group |
stat_centroid() | joint x and y summaries | geom_point(), geom_rug(), geom_margin_arrow(), etc. | data summary | group |
stat_summary_xy() | joint x and y summaries | geom_point(), geom_rug(), geom_margin_arrow(), etc. | data summary | group |
stat_functions() | compute y from x range | geom_line(), geom_point(), etc. | draw function curves | group |
Statistics defined in package ‘ggpp’.
Justification
Justifications "outward_mean", "inward_mean", "outward_median" and "inward_median" implement outward and inward justification relative to the centroid of the data instead of to the center of the $x$ or $y$ scales. Justification outward or inward from an arbitrary origin is also supported. Justification "position" implements justification at the edge nearest to the original position. This works only together with position functions that save the original location using the naming convention implemented in ‘ggpp’, otherwise default justification falls-back to "center"/"middle".
History
This package is a “spin-off” from package ‘ggpmisc’ containing extensions to the grammar originally written for use wihtin ‘ggpmisc’. As ‘ggpmisc’ had grown in size, splitting it into two packages was necessary to easy development and maintenance and to facilitate imports into other packages. For the time being, package ‘ggpmisc’ imports and re-exports all visible definitions from ‘ggpp’.
Examples
The plots below exemplify some of the things that ‘ggpp’ makes possible or makes easier to code compared to ‘ggplot’ used on its own. Additional examples including several combining ‘ggpp’ and ‘ggrepel’ are provided in the package vignette.
library(ggpp)
library(ggrepel)
library(dplyr)
Insets
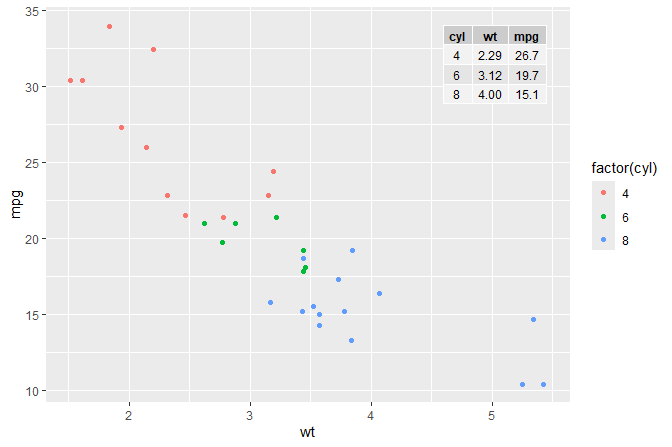
A plot with an inset table.
mtcars %>%
group_by(cyl) %>%
summarize(wt = mean(wt), mpg = mean(mpg)) %>%
ungroup() %>%
mutate(wt = sprintf("%.2f", wt),
mpg = sprintf("%.1f", mpg)) -> tb
df <- tibble(x = 5.45, y = 34, tb = list(tb))
ggplot(mtcars, aes(wt, mpg, colour = factor(cyl))) +
geom_point() +
geom_table(data = df, aes(x = x, y = y, label = tb))
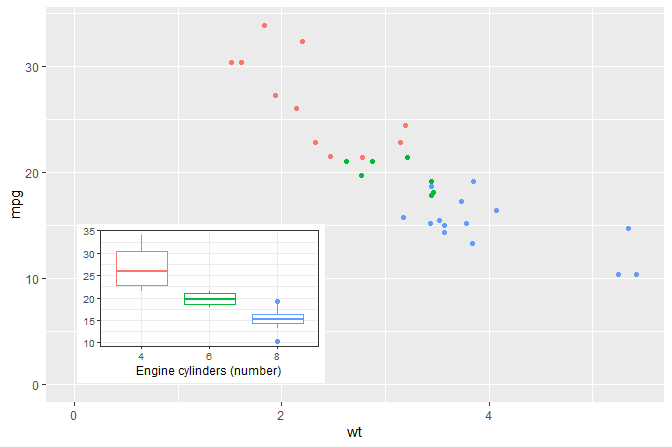
A plot with an inset plot.
Inset plot positioned using native plot coordinates (npc) using numerical values in the range 0..1 together with I().
p <- ggplot(mtcars, aes(factor(cyl), mpg, colour = factor(cyl))) +
stat_boxplot() +
labs(y = NULL, x = "Engine cylinders (number)") +
theme_bw(9) + theme(legend.position = "none")
ggplot(mtcars, aes(wt, mpg, colour = factor(cyl))) +
geom_point(show.legend = FALSE) +
annotate("plot", x = I(0.05), y = I(0.05), label = p,
hjust = "inward", vjust = "inward") +
expand_limits(y = 0, x = 0)
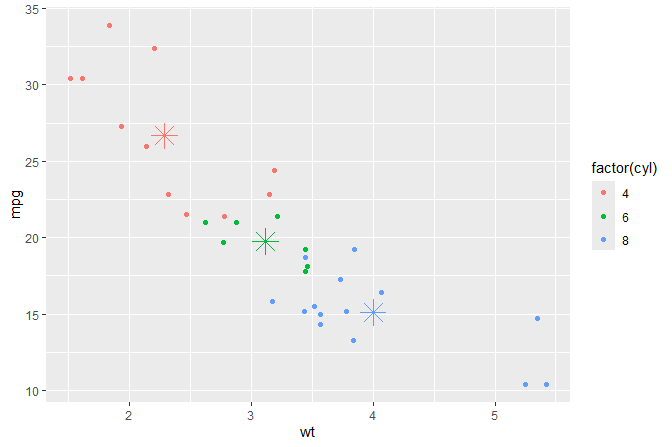
Centroids
Means computed on-the-fly and shown as asterisks.
ggplot(mtcars, aes(wt, mpg, colour = factor(cyl))) +
geom_point() +
stat_centroid(shape = "asterisk", size = 6)
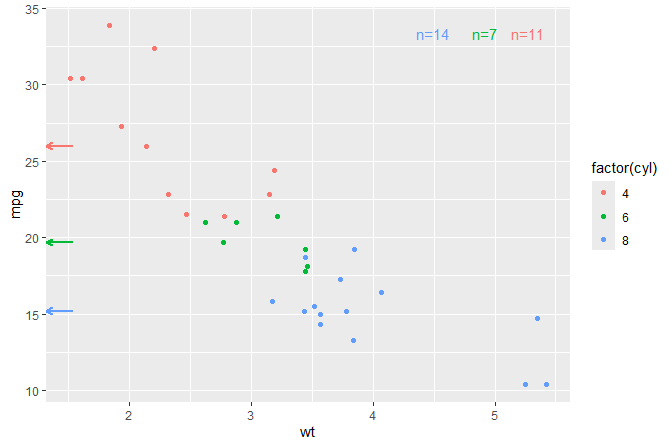
Medians computed on-the-fly shown as marginal arrows. Labels with number of observations per group.
ggplot(mtcars, aes(wt, mpg, colour = factor(cyl))) +
geom_point() +
stat_centroid(geom = "y_margin_arrow", .fun = median,
aes(yintercept = after_stat(y)), arrow.length = 0.05) +
stat_group_counts(vstep = 0, hstep = 0.09)
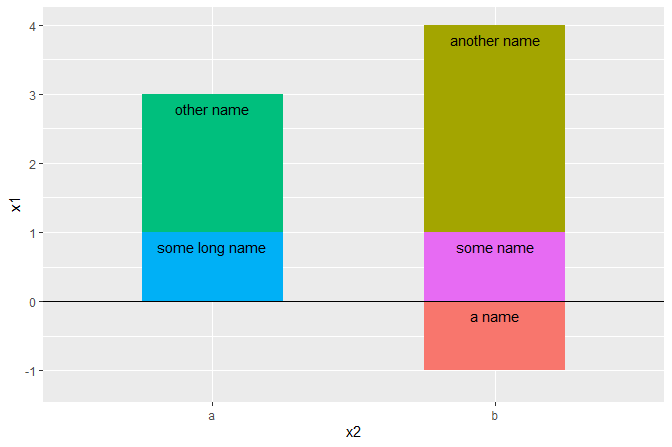
Nudging and stacking combined
df <- data.frame(x1 = c(1, 2, 1, 3, -1),
x2 = c("a", "a", "b", "b", "b"),
grp = c("some long name", "other name", "some name",
"another name", "a name"))
# Add labels to a horizontal column plot (stacked by default)
ggplot(data = df, aes(x2, x1, group = grp)) +
geom_col(aes(fill = grp), width=0.5) +
geom_hline(yintercept = 0) +
geom_text(
aes(label = grp),
position = position_stacknudge(vjust = 1, y = -0.2)) +
theme(legend.position = "none")
Installation
Installation of the most recent stable version from CRAN (sources, Mac and Win binaries):
install.packages("ggpp")
Installation of the current unstable version from R-Universe CRAN-like repository (binaries for Mac, Win, Webassembly, and Linux, as well as sources available):
install.packages('ggpp',
repos = c('https://aphalo.r-universe.dev',
'https://cloud.r-project.org'))
Installation of the current unstable version from GitHub (from sources):
# install.packages("devtools")
devtools::install_github("aphalo/ggpp")
Documentation
HTML documentation for the package, including help pages and the User Guide, is available at (https://docs.r4photobiology.info/ggpp/).
News about updates are posted at (https://www.r4photobiology.info/).
Chapter 7 in Aphalo (2020) and Chapter 9 in Aphalo (2024) explain basic concepts of the grammar of graphics as implemented in ‘ggplot2’ as well as extensions to this grammar including several of those made available by packages ‘ggpp’ and ‘ggpmisc’. Information related to the book is available at https://www.learnr-book.info/.
Contributing
Please report bugs and request new features at (https://github.com/aphalo/ggpp/issues). Pull requests are welcome at (https://github.com/aphalo/ggpp).
Citation
If you use this package to produce scientific or commercial publications, please cite according to:
citation("ggpp")
#> To cite package 'ggpp' in publications use:
#>
#> Aphalo P (2024). _ggpp: Grammar Extensions to 'ggplot2'_. R package
#> version 0.5.8, https://github.com/aphalo/ggpp,
#> <https://docs.r4photobiology.info/ggpp/>.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {ggpp: Grammar Extensions to 'ggplot2'},
#> author = {Pedro J. Aphalo},
#> year = {2024},
#> note = {R package version 0.5.8,
#> https://github.com/aphalo/ggpp},
#> url = {https://docs.r4photobiology.info/ggpp/},
#> }
Acknowledgements
Being an extension to package ‘ggplot2’, some of the code in package ‘ggpp’ has been created by using as a template that from layer functions and scales in ‘ggplot2’. The user interface of ‘ggpp’ aims at being as consistent as possible with ‘ggplot2’ and the layered grammar of graphics (Wickham 2010). New features added in ‘ggplot2’ are added when relevant to ‘ggpp’, such as support for orientation for flipping of layers. This package does consequently indirectly include significant contributions from several of the authors and maintainers of ‘ggplot2’, listed at (https://ggplot2.tidyverse.org/).
Coordination of development through a friendly exchange of ideas and reciprocal contributions by Kamil Slowikowski to ‘ggpp’ and by myself to ‘ggrepel’ has made the two packages fully inter-compatible.
References
Aphalo, Pedro J. (2024) Learn R: As a Language. 2ed. The R Series. Boca Raton and London: Chapman and Hall/CRC Press. ISBN: 9781032516998. 466 pp.
Aphalo, Pedro J. (2020) Learn R: As a Language. 1ed. The R Series. Boca Raton and London: Chapman and Hall/CRC Press. ISBN: 9780367182533. 350 pp.
Wickham, Hadley. 2010. “A Layered Grammar of Graphics.” Journal of Computational and Graphical Statistics 19 (1): 3–28. https://doi.org/10.1198/jcgs.2009.07098.
License
© 2016-2024 Pedro J. Aphalo ([email protected]). Released under the GPL, version 2 or greater. This software carries no warranty of any kind.

