Description
Post-Process 'ggplot2' Plots with 'TikZ' Code Using Plot Coordinates.
Description
Annotation of 'ggplot2' plots with arbitrary 'TikZ' code, using absolute data or relative plot coordinates.
README.md
ggtikz
ggtikz allows you to annotate plots created using ggplot2 with arbitrary TikZ code when rendering them with the tikzDevice. The annotations can be made using data coordinates, or with coordinates relative to a specified panel or the whole plot.
Plots with multiple panels (via facet_grid() or facet_wrap()) are supported.
For a few examples, see the examples vignette.
Installation
You can install the latest ggtikz release from CRAN with:
install.packages("ggtikz")
Or get the development version from GitHub:
# install.packages("devtools")
devtools::install_github("osthomas/ggtikz", ref = "devel")
Basic Usage
- Create a ggplot.
- Add annotations with
ggtikz().
library(ggplot2)
library(ggtikz)
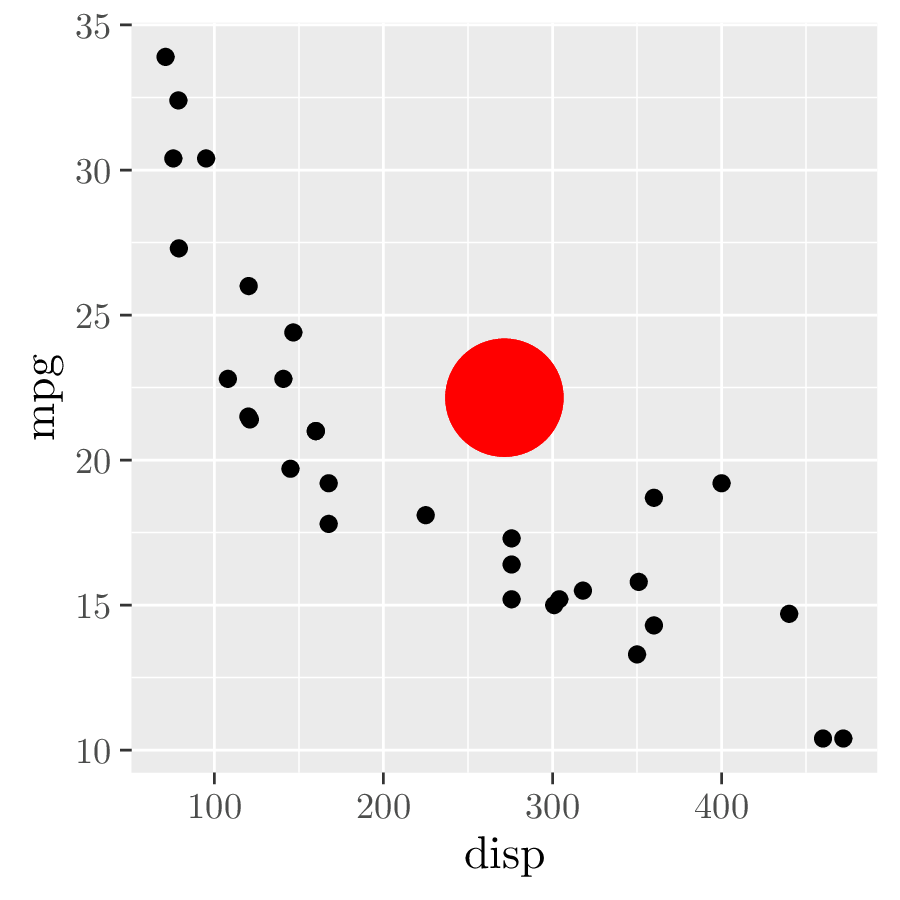
p <- ggplot(mtcars, aes(disp, mpg)) + geom_point() # 1.
## tikz("plot.tikz")
ggtikz(p,
"\\fill[red] (0.5,0.5) circle (5mm);",
xy = "panel", panelx = 1, panely = 1)
## dev.off()
## Render with LaTeX ...
Advanced Usage
- Create a ggplot.
- Create a ggtikz canvas from the plot with
ggtikzCanvas(). - Create ggtikz annotations with
ggtikzAnnotation(). - Add the annotations to the canvas.
- Draw the plot and the annotations using tikzDevice.
library(ggplot2)
library(ggtikz)
p <- ggplot(mtcars, aes(disp, mpg)) + geom_point() # 1.
canvas <- ggtikzCanvas(p) # 2.
annot1 <- ggtikzAnnotation( # 3.
"
\\draw (0,0) -- (1,1);
\\draw (0,1) -- (1,0);
\\fill[red] (0.5,0.5) circle (5mm);
",
xy = "panel", panelx = 1, panely = 1
)
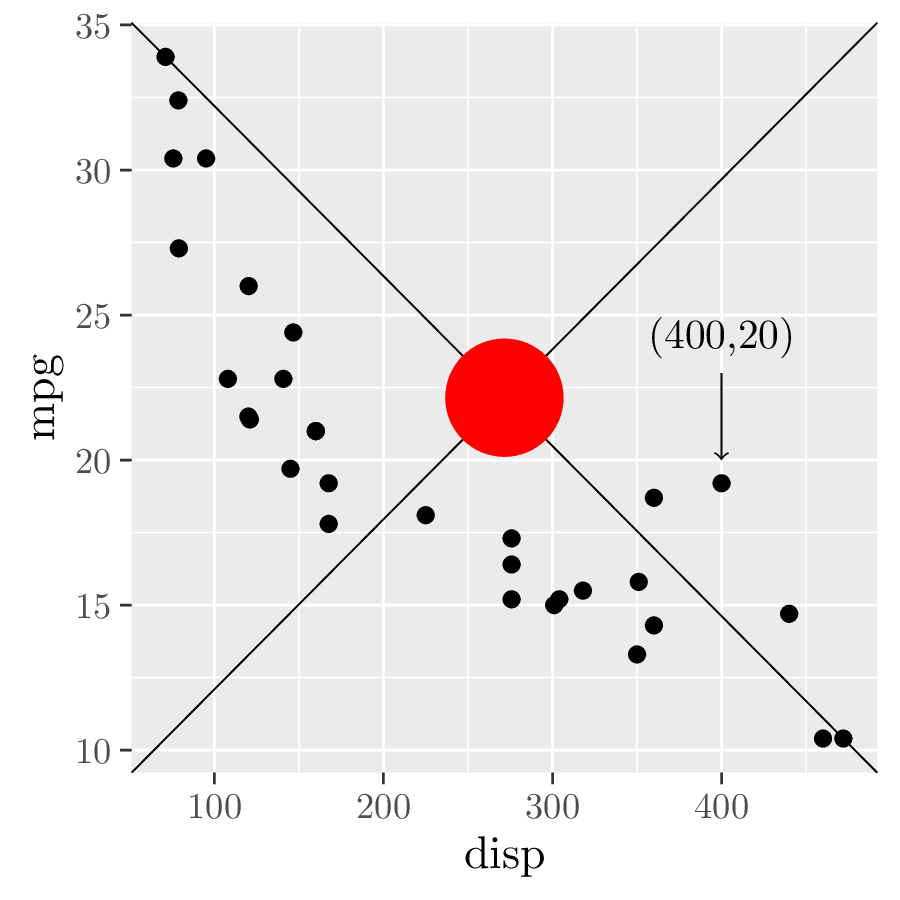
annot2 <- ggtikzAnnotation( # 3.
"\\draw[<-] (400,20) -- ++(0,3) node[at end, anchor=south] {(400,20)};",
xy = "data", panelx = 1, panely = 1
)
## tikz("plot.tikz")
p # 4. + 5.
canvas + annot1 + annot2
## dev.off()
## Render with LaTeX ...