Real-Time PCR Data Sets by Guescini et al. (2008).
guescini
{guescini} is an R data package that provides real-time PCR raw fluorescence data by Guescini et al. (2008) in tidy format.
Installation
Install {guescini} from CRAN:
# Install from CRAN
install.packages("guescini")
Data
Guescini et al. (2008) explored the effect of amplification inhibition on qPCR quantification. Two systems were devised to alter the amplification efficiency:
- decreasing of the amplification mix used in the reaction
- increasing of IgG (PCR inhibitor) concentration in the reaction
The raw fluorescence data associated with the decreasing of the amplification mix is provided as the data set amp_mix_perc; the data obtained with increasing concentrations of IgG is provided as IgG_inhibition.
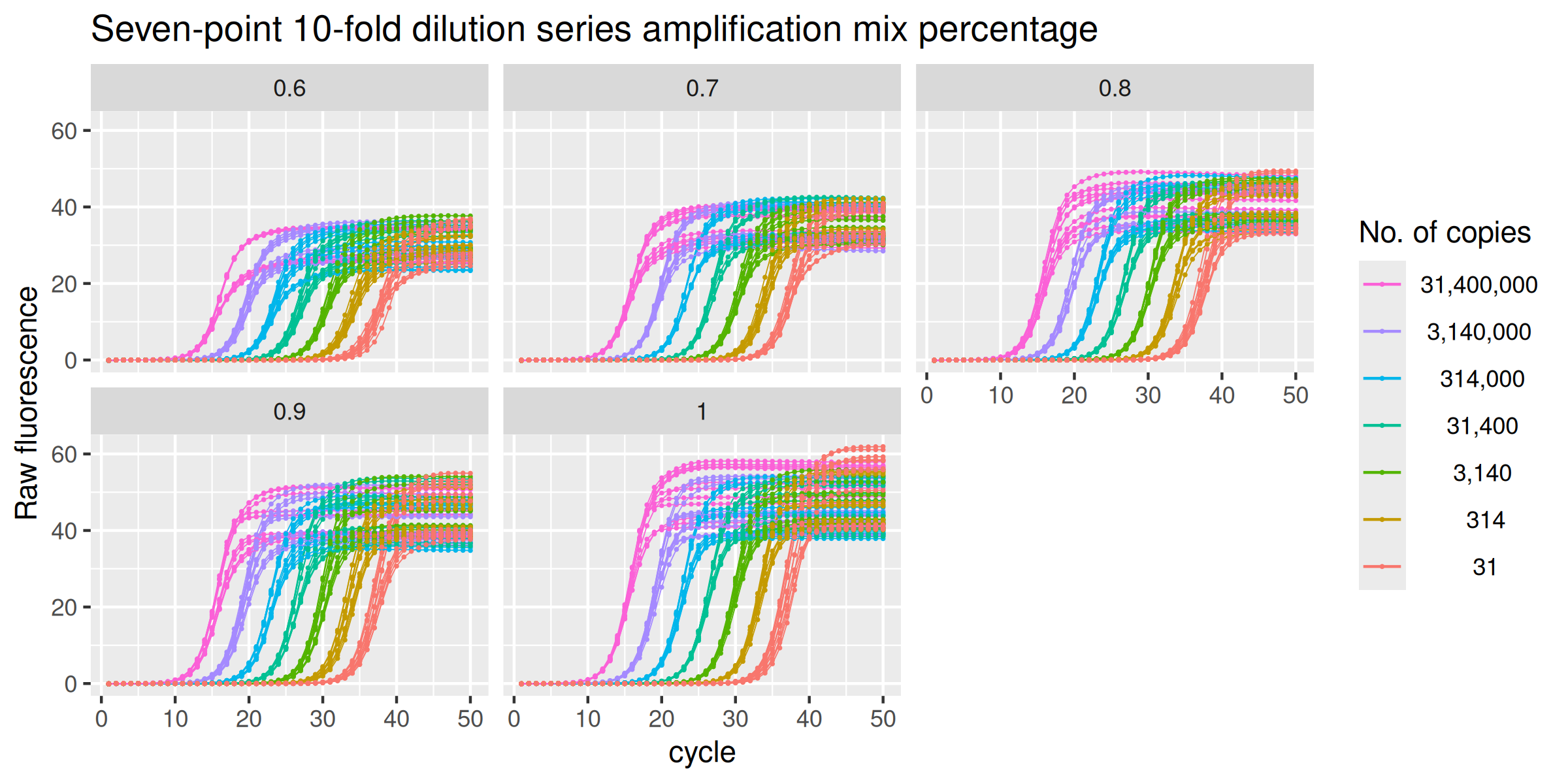
Amplification mix percentage
The data set amp_mix_perc corresponds to a set of amplification runs where the MT-ND1 gene is amplified in reactions having the same initial amount of DNA but different amounts of SYBR Green I Master mix. A standard curve was performed over a wide range of input DNA ($3.14 \times 10^7\ \text{thru}\ 3.14 \times 10^1$) in the presence of optimal amplification conditions (100% amplification mix), while the unknowns were run in the presence of the same starting DNA amounts but with amplification mix quantities ranging from 60% to 100%.
library(guescini)
amp_mix_perc
#> # A tibble: 21,000 × 12
#> plate well dye target sample_type run replicate amp_mix_perc copies
#> <fct> <fct> <fct> <fct> <fct> <fct> <fct> <dbl> <int>
#> 1 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 2 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 3 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 4 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 5 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 6 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 7 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 8 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 9 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 10 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> # ℹ 20,990 more rows
#> # ℹ 3 more variables: dilution <int>, cycle <int>, fluor <dbl>
amp_mix_perc %>%
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(run, amp_mix_perc, copies),
col = format(copies, big.mark = ",", scientific = FALSE)
)) +
geom_line(linewidth = 0.2) +
geom_point(size = 0.2) +
labs(y = "Raw fluorescence", colour = "No. of copies", title = "Seven-point 10-fold dilution series amplification mix percentage") +
guides(color = guide_legend(override.aes = list(linewidth = 0.5), reverse = TRUE)) +
facet_wrap(vars(amp_mix_perc))
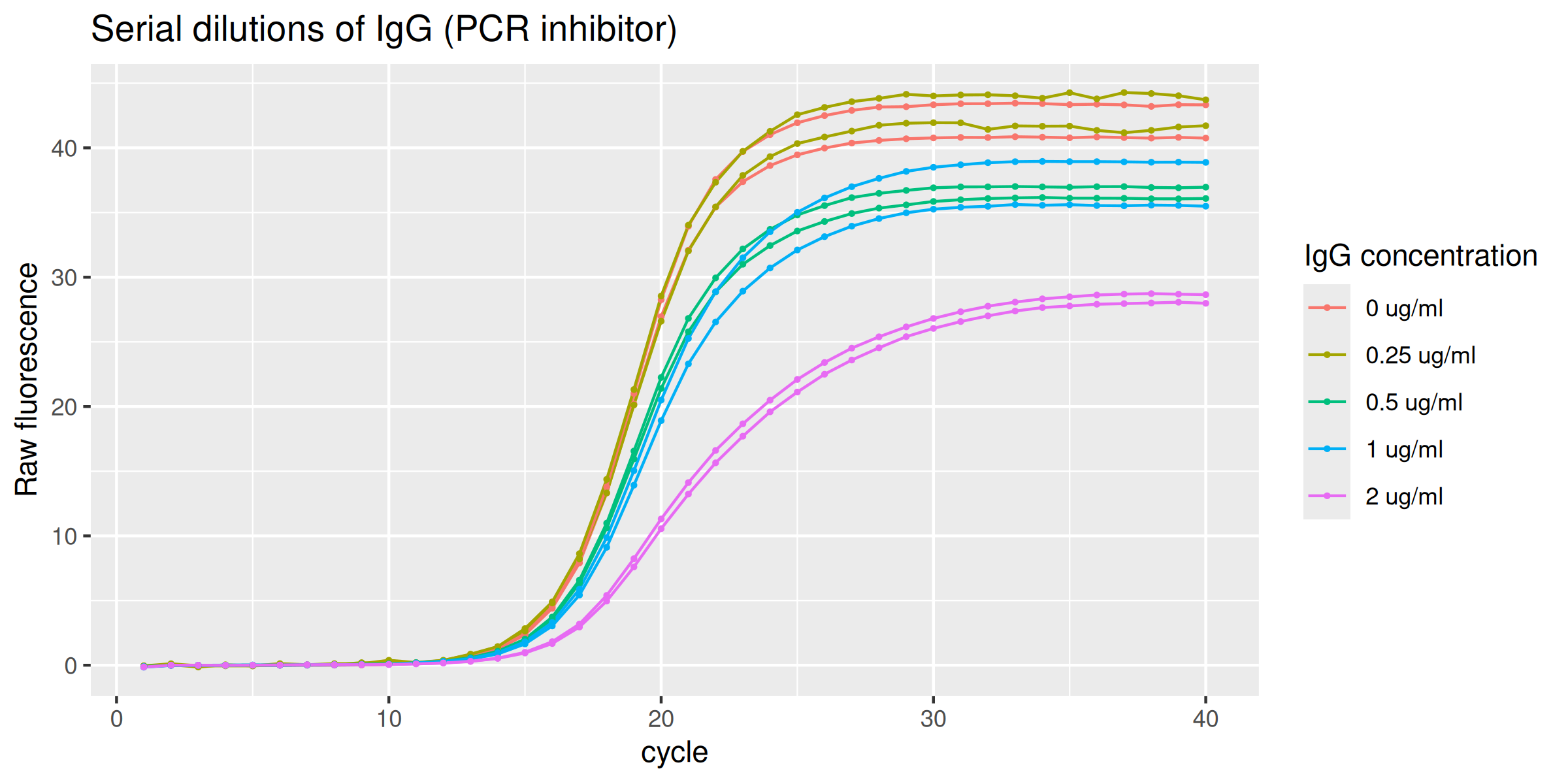
Inhibition by IgG
The data set IgG_inhibition provides those runs performed in the presence of an optimal amplification reaction mix added with serial dilutions of IgG (0.0 - 2 ug/ml) thus acting as the inhibitory agent.
IgG_inhibition %>%
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(IgG_conc, replicate),
col = paste(as.character(IgG_conc), "ug/ml")
)) +
geom_line(linewidth = 0.5) +
geom_point(size = 0.5) +
labs(y = "Raw fluorescence", colour = "IgG concentration", title = "Serial dilutions of IgG (PCR inhibitor)") +
guides(color = guide_legend(override.aes = list(linewidth = 0.5)))
Code of Conduct
Please note that the guescini project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
References
Michele Guescini, Davide Sisti, Marco BL Rocchi, Laura Stocchi and Vilberto Stocchi. A new real-time PCR method to overcome significant quantitative inaccuracy due to slight amplification inhibition. BMC Bioinformatics 9:326 (2008). doi: 10.1186/1471-2105-9-326.