Description
Make Forest Plot with GWAS Data.
Description
Extract and reform data from GWAS (genome-wide association study) results, and then make a single integrated forest plot containing multiple windows of which each shows the result of individual SNPs (or other items of interest).
README.md
gwasforest
The goal of gwasforest is to extract and reform data from GWAS results, and then make a single integrated forest plot containing multiple windows of which each shows the result of individual SNPs (or other items of interest).
Installation
The development version of gwasforest can be installed from GitHub with:
devtools::install_github("yilixu/gwasforest", ref = "main")
Quick Demos
- ( 1 ) when customFilename (main input) is in dataframe format (with standardized column names):
library(gwasforest)
set.seed(123)
# generate example data
tempValue = runif(n = 18, min = 0.01, max = 2)
tempStdErr = tempValue / rep(3:5, times = 6)
eg_customFilename = data.frame(paste0("Marker", 1:6), tempValue[1:6], tempStdErr[1:6], tempValue[7:12], tempStdErr[7:12], tempValue[13:18], tempStdErr[13:18], stringsAsFactors = FALSE)
colnames(eg_customFilename) = c("MarkerName", paste0(rep("Study", times = 6), rep(1:3, each = 2), rep(c("__Value", "__StdErr"), times = 3)))
rm(tempValue, tempStdErr)
# run gwasforest function
eg_returnList = gwasforest(eg_customFilename, stdColnames = TRUE, valueFormat = "Effect", metaStudy = "Study1", colorMode = "duo")
#> [1] "Column names are in the same format as instruction example"
#> [1] "Loading user-provided values"
#> [1] "Start calculating Confidence Interval (non-exponential)"
#> [1] "Based on user's choice, GWAS results output file will not be generated"
#> [1] "All studies except meta study will be set in alphabetical order from top to bottom on the forest plot"
#> Registered S3 methods overwritten by 'ggplot2':
#> method from
#> [.quosures rlang
#> c.quosures rlang
#> print.quosures rlang
#> [1] "Based on user's choice, GWAS forest plot file will not be generated"
#> [1] "Run completed, thank you for using gwasforest"
- ( 2 ) when customFilename is in dataframe format (without standardized column names), while customFilename_studyName is provided in dataframe format:
# generate example data
tempValue = runif(n = 18, min = 0.01, max = 2)
tempStdErr = tempValue / rep(3:5, times = 6)
eg_customFilename = data.frame(paste0("Marker", 1:6), tempValue[1:6], tempStdErr[1:6], tempValue[7:12], tempStdErr[7:12], tempValue[13:18], tempStdErr[13:18], stringsAsFactors = FALSE)
colnames(eg_customFilename) = c("MarkerName", paste0(rep("Study", times = 6), rep(1:3, each = 2), sample(LETTERS, 6)))
rm(tempValue, tempStdErr)
eg_customFilename_studyName = data.frame("studyName" = paste0("Study", 1:3), stringsAsFactors = FALSE)
# run gwasforest function
eg_returnList = gwasforest(eg_customFilename, customFilename_studyName = eg_customFilename_studyName, stdColnames = FALSE, customColnames = c("Value", "StdErr"), valueFormat = "Effect", metaStudy = "Study1", colorMode = "duo")
- ( 3 ) when customFilename_results is in dataframe format (run either of the two steps above to see the example results):
# extract results table
eg_customFilename_results = eg_returnList[[1]]
library(ggplot2)
# render plot, see additional NOTES below
plot(eg_returnList[[2]])
#> Warning: Removed 12 rows containing missing values (geom_text_repel).
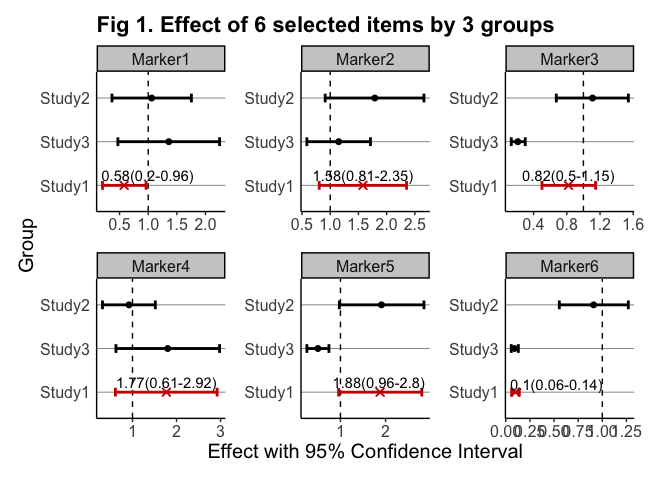
- ( 4 ) NOTES: As shown above, the plot rendered through plot() may suffer from certain issues such as low-resolution and overlapping labels. To overcome these issues and get the genuine plot output from gwasforest, it is recommended to provide a valid outputFolderPath so that a better-rendered plot can be created. The below is the genuine plot output created from the same example data: