Wrangle Phylogenetic Distance Matrices and Other Utilities.
harrietr: An R package for various phylogenetic and evolutionary biology data manipulations
Why harrietr:
Harriet is believed to be Charles Darwin's pet giant turtle. It is thought that Harriet spent the latter part of her life at in Brisbane, Australia. Thus, Harriet satisfies the three criteria to name this package: (1) it is somehow evolutionarily related; (2) it has an Australian connection; and (3) it avoids the prefix phylo used by many R evolutionarily- relavant packages. The appended r just helps differentiate to make it easier to search in Google, and aludes to the fact that it is related to the programming language R.
How to get it
From CRAN:
Add
Bioconductorto your list of default repositories:setRepositories(ind = 1:2)Install
harrietr:install.packages("harrietr", dependencies = TRUE)
Latest and gratest version from GitHub:
You must use devtools:
If you don't have
devtoolsinstalled:install.packages('devtools')Add
Bioconductorto your list of default repositories:setRepositories(ind = 1:2)Install
harrietr:devtools::install_github("andersgs/harrietr")
How to use it
Three functions are provided at this time:
dist_long --- This function takes as input an alignment in DNA.bin format calculates all the pairwise distances, and returns all the unique pairwise distances as a data.frame in a long format. For example, in the case of three samples:
| id1 | id2 | distance |
|---|---|---|
| sample1 | sample2 | dist_12 |
| sample1 | sample3 | dist_13 |
| sample2 | sample3 | dist_23 |
You can give dist_long a tree (object of class phylo), and it will add a fourth column with the pairwise distance obtained from the tree:
| id1 | id2 | distance | evol_dist |
|---|---|---|---|
| sample1 | sample2 | dist_12 | evol_dist_12 |
| sample1 | sample3 | dist_13 | evol_dist_13 |
| sample2 | sample3 | dist_23 | evol_dist_23 |
melt_dist --- This function is used by dist_long, but it takes as input a distance matrix. This might be useful if you alredy have a distance matrix that is imported into R.
get_node_support --- This function is written to work with trees generated by IQTREE. In particular, if the tree was generated when calculating node support by both ultrafast bootstrap and SH approximate likelihood ratio test, IQTREE writes the support as the node label in the Newick file in the following format: "SH-aLRT/uBS". In other words, it is a string with two values separated by a slash. The first value is the SH-aLRT support (as a percentage) and the second value is the ultrafast bootstrap support (also as a percentage).
The output is a data.frame with each row representing an internal node, with information that can be used to plot support information layers on a tree.
Some use cases
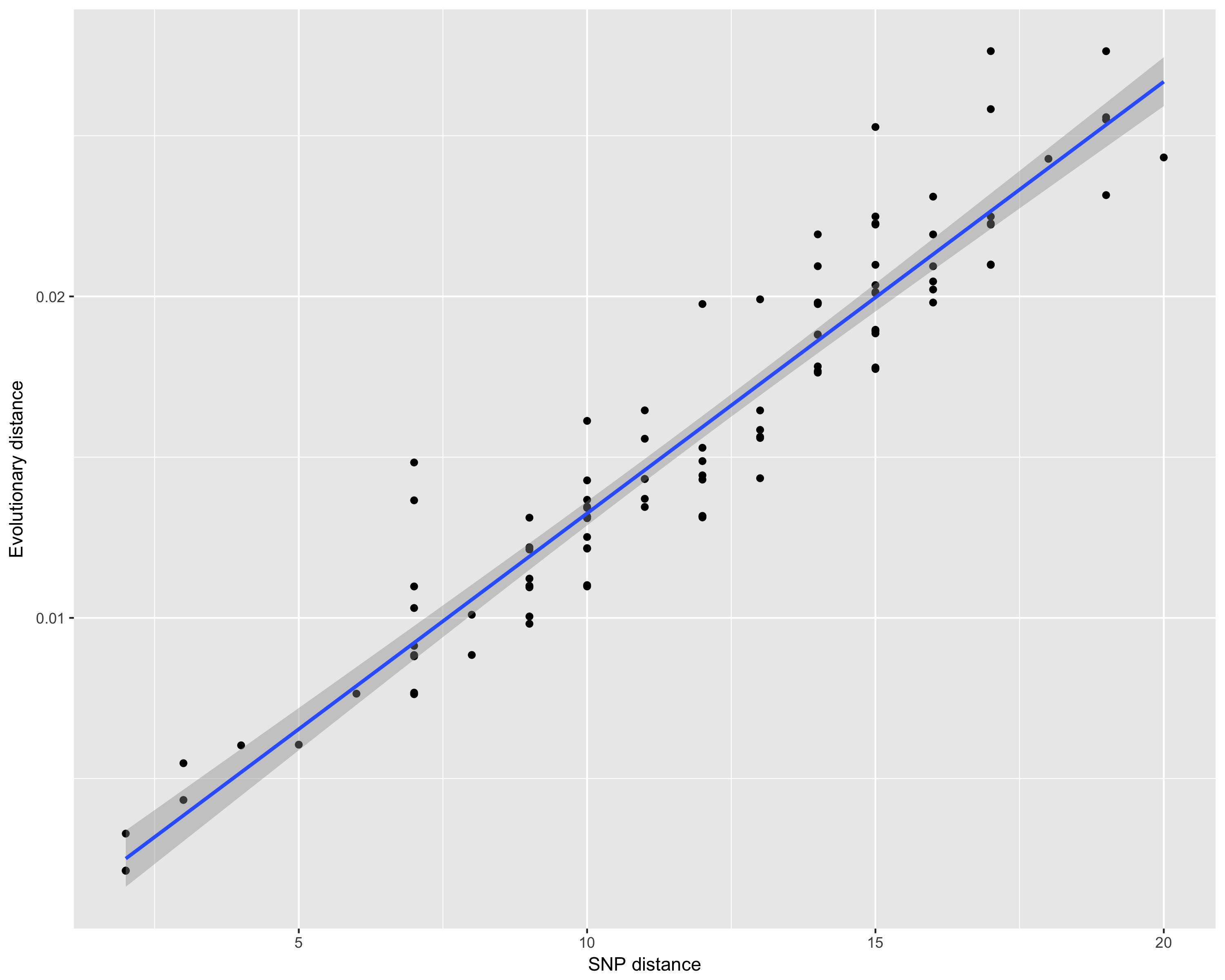
Comparing distances
Assume you have a tree, and you want to understand what is the relationship between the branch lengths and the number of SNPs. The function dist_long can help you get there:
library(harrietr)
library(ggplot2)
data("woodmouse")
data("woodmouse_iqtree")
dist_df <- dist_long(aln = woodmouse, order = woodmouse_iqtree$tip.label, dist = "N", tree = woodmouse_iqtree)
ggplot(dist_df, aes(x = dist, y = evol_dist)) +
geom_point() + stat_smooth(method = 'lm') +
ylab("Evolutionary distance") +
xlab("SNP distance")
This will produce the following image:
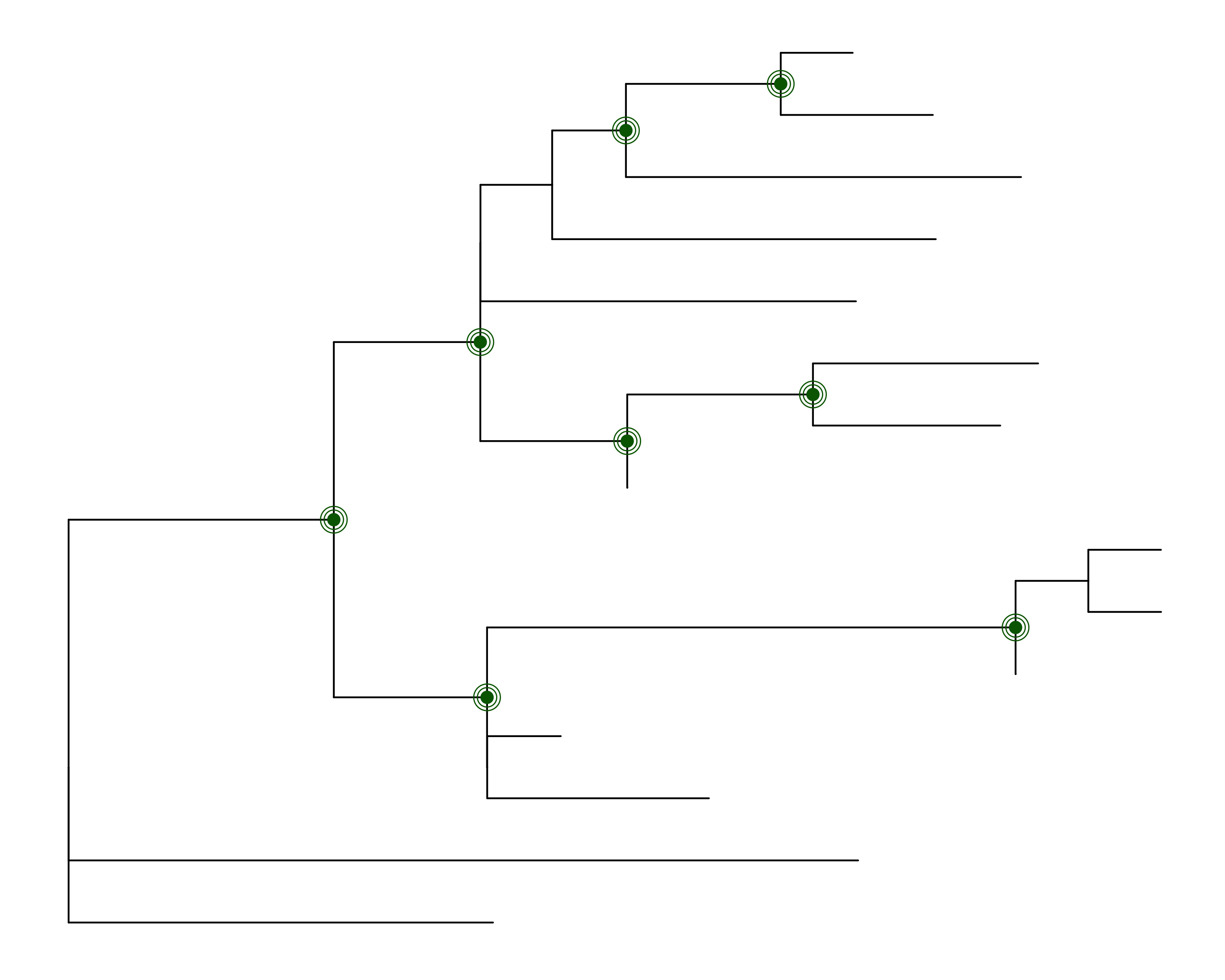
Indicating nodes that have support on a tree
Assume you have generated your ML tree with IQTREE, and wish to plot it in R, and indicate which nodes have 50% or more support values for both metrics (note: the value of 50% is likely too low, these values are chosen only for illustration purposes). The function get_node_support can help you get there:
library(ggtree)
library(dplyr)
library(harrietr)
data("woodmouse_iqtree")
p1 <- ggtree(woodmouse_iqtree)
node_support <- get_node_support(woodmouse_iqtree)
p1 +
geom_point(data = node_support %>% dplyr::filter(`SH-aLRT` >= 50 & uBS >= 50), aes(x = x, y = y), colour = 'darkgreen', size = 3) +
geom_point(data = node_support %>% dplyr::filter(`SH-aLRT` >= 50 & uBS >= 50), aes(x = x, y = y), colour = 'darkgreen', size = 5, pch = 21) +
geom_point(data = node_support %>% dplyr::filter(`SH-aLRT` >= 50 & uBS >= 50), aes(x = x, y = y), colour = 'darkgreen', size = 7, pch = 21)
This will produce the following image:
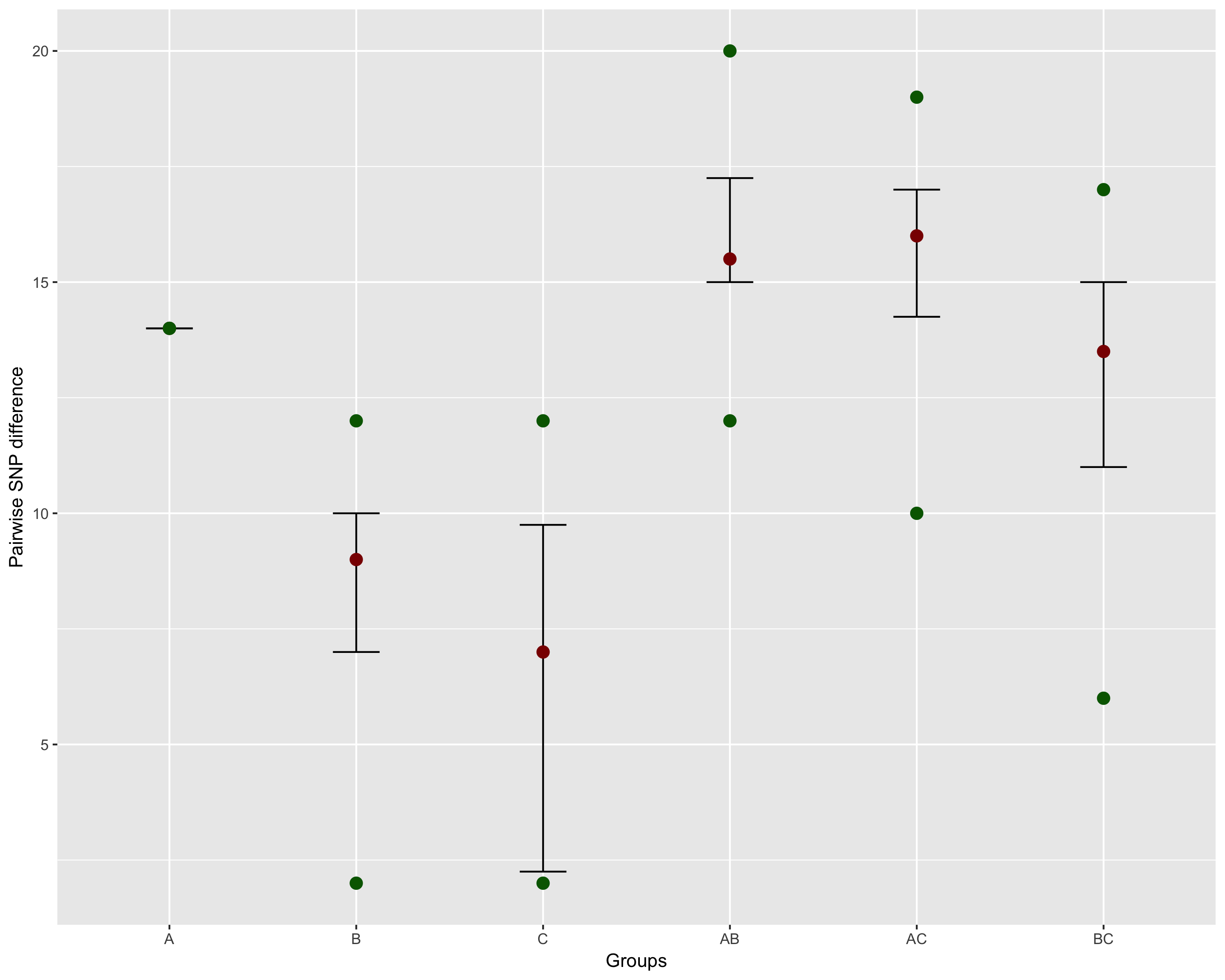
Getting group level statistics
Assume you have classified your samples into different groups (say A, B, and C). These could be anything (e.g., MLST, sample source, host, etc.), and you want summary information among and between the groups (e.g., IQR, min/max dist). You can use dist_long and add_metadata to generate the data.frame you need:
library(ggplot2)
library(dplyr)
library(harrietr)
data("woodmouse")
data("woodmouse_iqtree")
data("woodmouse_meta")
dist_df <- dist_long(aln = woodmouse, order = woodmouse_iqtree$tip.label, dist = "N", tree = woodmouse_iqtree)
dist_df <- add_metadata(dist_df, woodmouse_meta, isolate = 'SAMPLE_ID', group = 'CLUSTER', remove_ind = TRUE)
dist_df %>%
dplyr::group_by(CLUSTER) %>%
dplyr::summarise(q50 = median(dist),
q25 = quantile(dist, prob = c(0.25)),
q75 = quantile(dist, prob = c(0.75)),
min_dist = min(dist),
max_dist = max(dist)) %>%
ggplot( aes( x = CLUSTER, y = q50)) +
geom_errorbar( aes(ymin = q25, ymax = q75),width = 0.25 ) +
geom_point(size = 3, colour = 'darkred') +
geom_point( aes( y = min_dist), colour = 'darkgreen', size = 3) +
geom_point( aes( y = max_dist), colour = 'darkgreen', size = 3) +
ylab("Pairwise SNP difference") +
xlab("Groups")
This will produce the following image: