Description
Simulating the Development of h-Index Values.
Description
H-index and h-alpha are a bibliometric indicators. This package provides functions to simulate how these indicators may develop over time for a given set of researchers and to visualize the simulation data. The implementation is based on the 'STATA' ado h-index and is described in more detail in Bornmann et al. (2019) <arXiv:1905.11052>.
README.md
hindex
This package provides functionality to simulate the development of h-index and h-alpha values of scientists who collaborate on writing papers and to visualize the simulated data. The effect of publishing, being cited, and strategic collaborating can be simulated. The implementation is based on the STATA ado h-index.
Installation
Install the package from CRAN:
install.packages('hindex')
Install the latest version from GitHub using the devtools package:
# if devtools is not installed yet:
install.packages("devtools")
devtools::install_github("atekles/hindex")
Example
library(hindex)
set.seed(1234)
simdata <- simulate_hindex(runs = 5, n = 200, periods = 20, coauthors = 3, distr_initial_papers = 'poisson', dpapers_pois_lambda = 10, distr_citations = 'poisson', dcitations_mean = 5, dcitations_peak = 3, alpha_share = .33)
#> run 1...
#> run 2...
#> run 3...
#> run 4...
#> run 5...
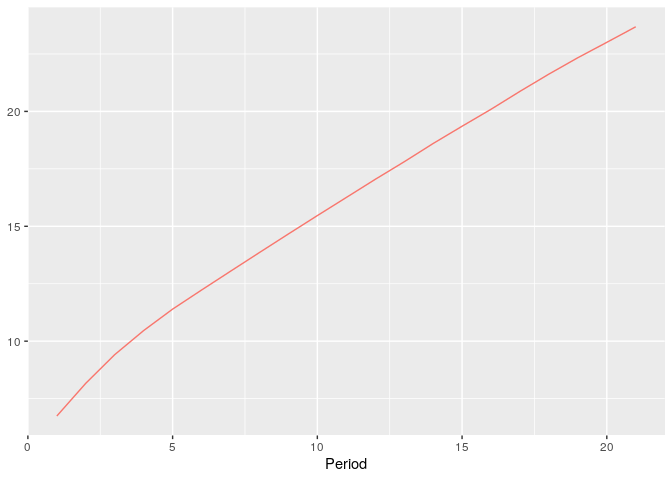
plot_hsim(simdata, plot_hindex = TRUE)
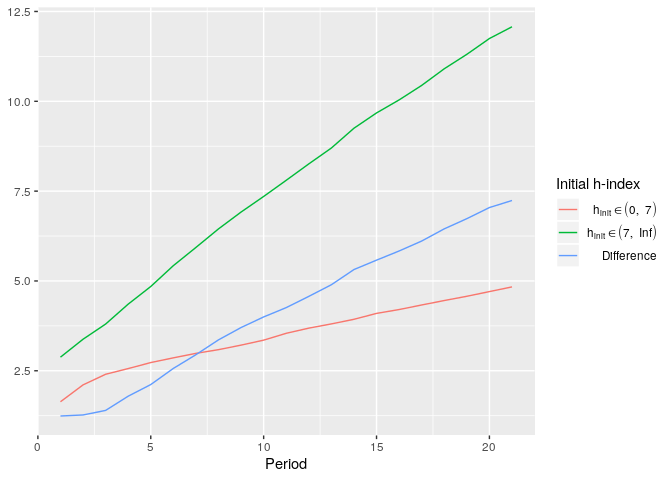
plot_hsim(simdata, plot_halpha = TRUE, group_boundaries = 'median', exclude_group_boundaries = TRUE, plot_group_diffs = TRUE)
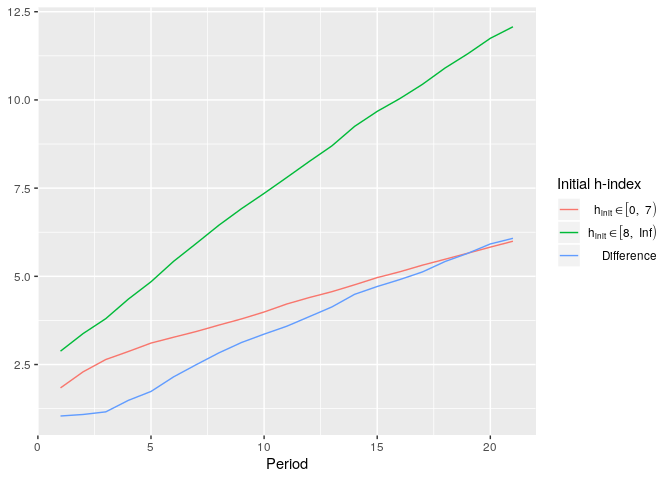
plot_hsim(simdata, plot_halpha = TRUE, group_boundaries = list(c(0, 7), c(8, Inf)), plot_group_diffs = TRUE)
simdata <- simulate_hindex(runs = 5, n = 200, periods = 20, subgroups_distr = .5, coauthors = 3, distr_initial_papers = 'poisson', dpapers_pois_lambda = 10, distr_citations = 'poisson', dcitations_mean = 5, dcitations_peak = 3, alpha_share = .33)
#> run 1...
#> run 2...
#> run 3...
#> run 4...
#> run 5...
plot_hsim(simdata, plot_hindex = TRUE, subgroups = TRUE, plot_group_diffs = TRUE)
