A Tool Used to Conduct Hypsometric Analysis of a Watershed.
hypsoLoop
hypsoLoop has been developed as tool to automate the drawing of hypsographic curves and the calculation of hypsometric curves for a watershed that contains several sub-catchments.
Installation
You can install the development version of hypsoLoop from GitHub with:
# devtools::install_github("fgashakamba/hypsoLoop")
and you can install the published beta version at CRAN with:
# Still in beta stage (version 0.2.0)
# Package currently under review by the CRAN team and it'll Soon be available at:
install.packages("hypsoLoop")
Example use of hypsoLoop
The workhorse function of hypsoLoop is called drawHypsocurves(). It accepts two arguments: a watersheds boundary layer (an sf or sp object) and an digital elevation model (DEM) covering the area of interest (a rasterLayer object). Additionally, the user may specify whether they want to get an output folder created in the root directory or not. If not, the hypsometric curves are drawn in the viewer and a summary table containing the hypsometric integers and other useful information on the sub-catchments is returned.
library(hypsoLoop)
drawHypsoCurves(watersheds, DEM)
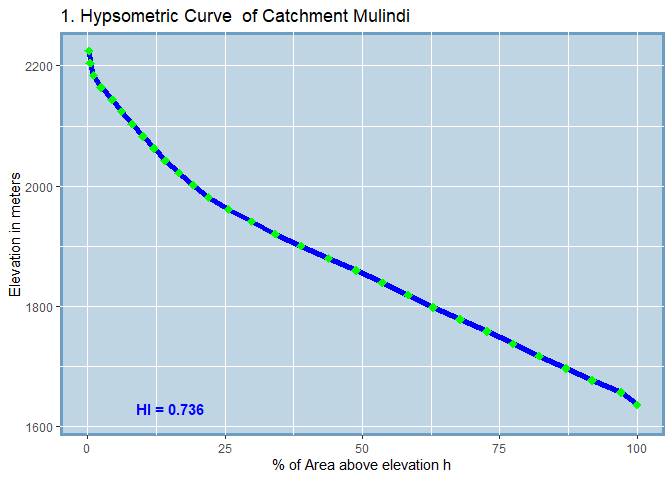
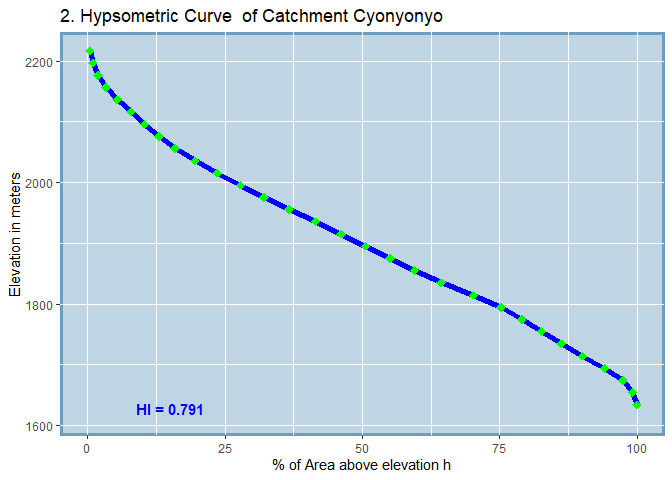
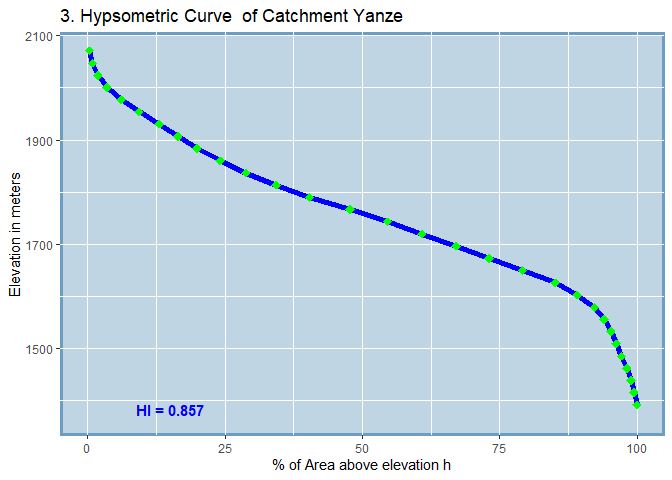
#> SUB_CATCHMENT_CODE MIN_ELEV MAX_ELEV TOTAL_AREA H_INTEGRAL
#> 1 Mulindi 1616 2225 2576.38 0.736
#> 2 Cyonyonyo 1614 2217 3673.1 0.791
#> 3 Yanze 1368 2071 3435.65 0.857
Other two functions are exposed from this hypsoLoop. These are the generateHypsoTables() which can be used to generate area-elevation tables for the sub-catchments of a watersheds and whose input requirements are the same as drawHypsoCurves(). The calc_areas() function is a handy function that be used to calculate the areas covered by each class of a categorical raster. It accepts one input argument which has to be a categorical raster of type rasterLayer and returns a table that summarizes the areas (in Hectares) covered by each class or category defined in the input raster.
library(hypsoLoop)
calc_areas(lulcYanze)
#> CLASS AREA
#> 1 1 1375.4506114
#> 2 2 528.6022953
#> 3 3 6830.5717454
#> 4 5 93.0439997
#> 5 6 0.2003449
#> 6 10 422.4313268
#> 7 11 401.4568430