INsulin Secretion ANalysEr.
INsulin Secretion ANalysEr 
A user-friendly interface, using Shiny, to analyse glucose-stimulated insulin secretion (GSIS) assays in pancreatic beta cells or islets.
The package allows the user to import several sets of experiments from different spreadsheets and to perform subsequent steps: summarise in a tidy format, visualise data quality and compare experimental conditions without omitting to account for technical confounders such as the date of the experiment or the technician.
Together, insane is a comprehensive method that optimises pre-processing and analyses of GSIS experiments in a friendly-user interface.
The Shiny App was initially designed for EndoC-betaH1 cell line following method described in Ndiaye et al., 2017 (https://doi.org/10.1016/j.molmet.2017.03.011).
Installation
# Install insane from CRAN:
install.packages("insane")
# Or the the development version from GitHub:
# install.packages("remotes")
remotes::install_github("mcanouil/insane")
library("insane")
go_insane()
Overview
The Shiny (R package) application insane (INsulin Secretion ANalysEr) provides a web interactive tool to import experiments of insulin secretion using cell lines such as EndoC-βH1.
Excel Template (top)
An Excel template is provided within the app to help users import their experiments in an easy way.
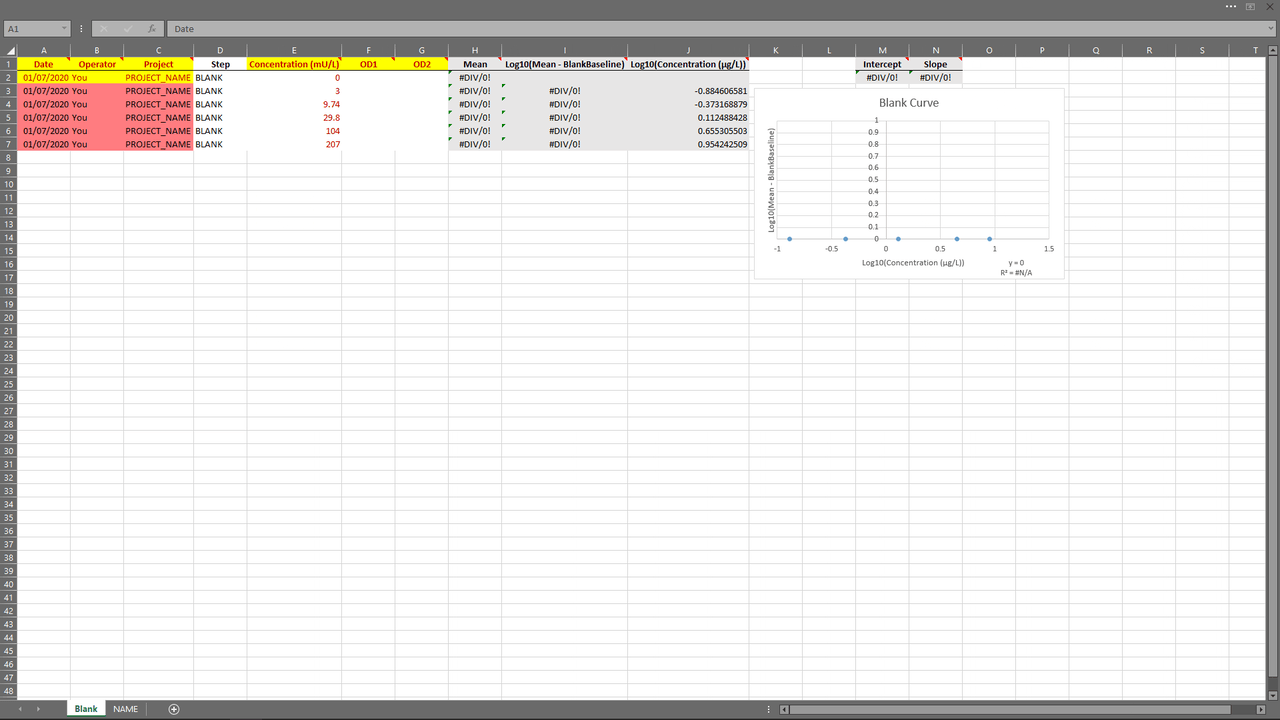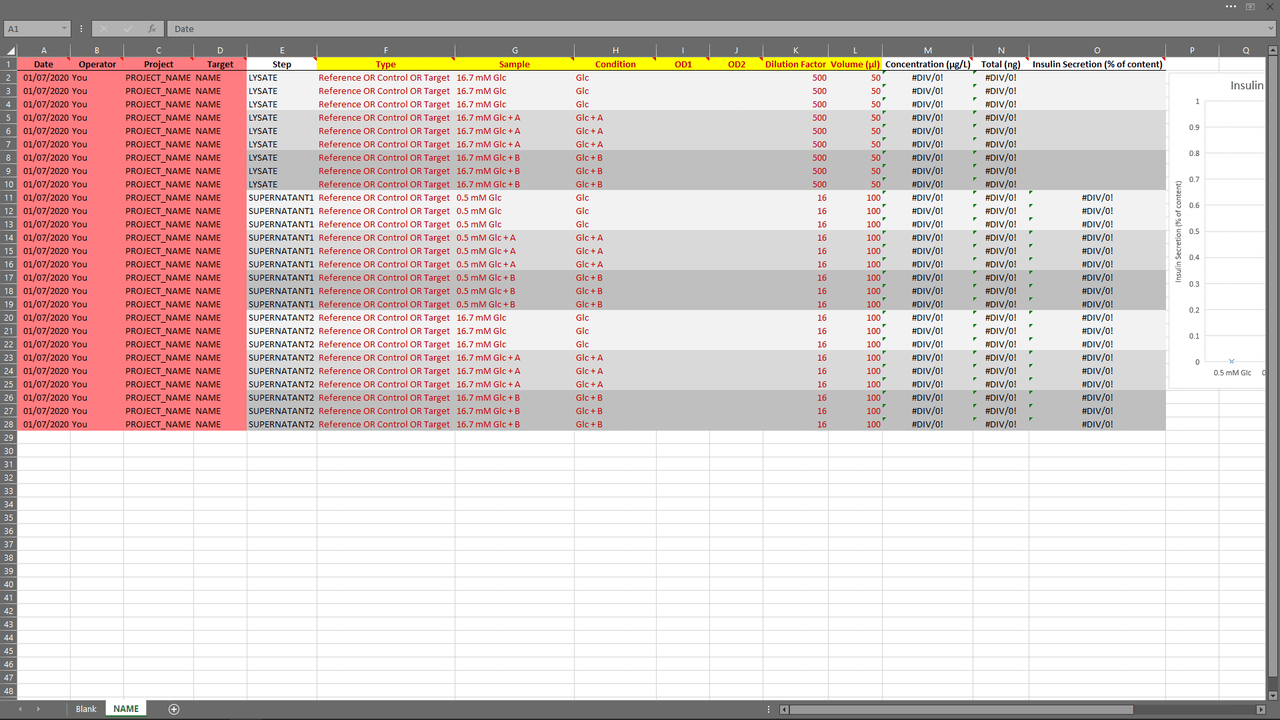
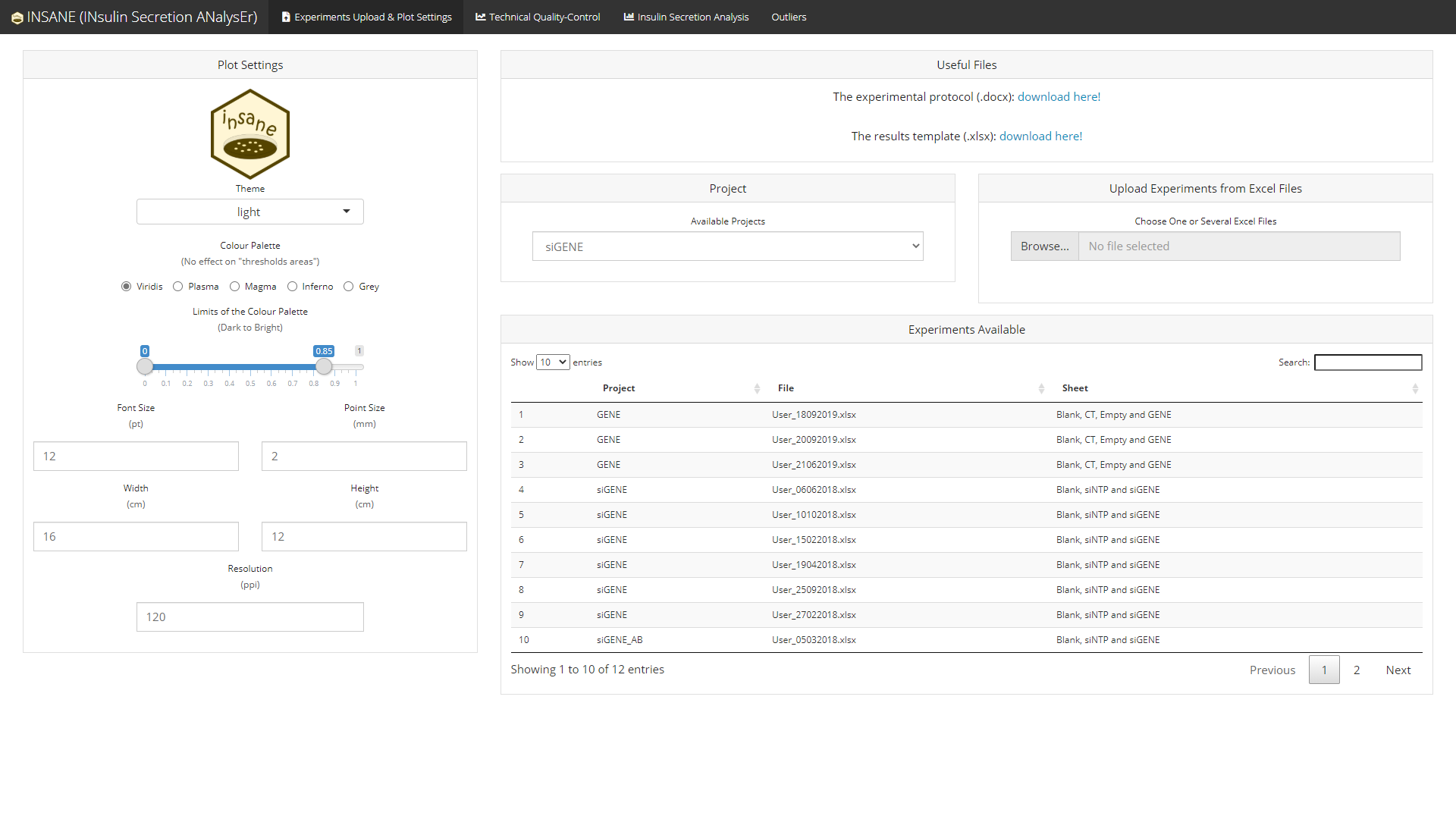
The App (top)
insane provides a user-friendly interface which can handle several projects separately.
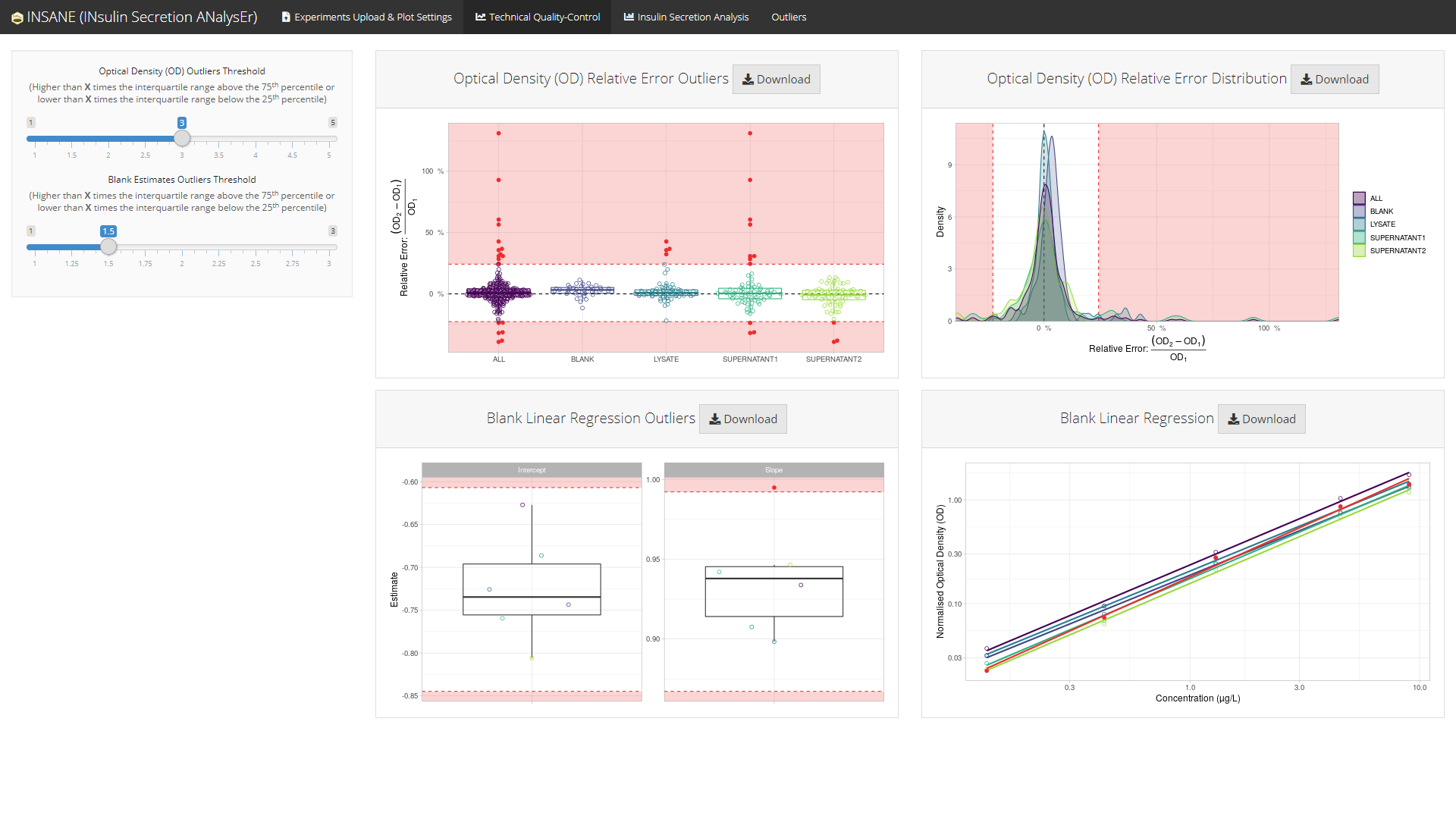
Technical Quality-Control (top)
insane performs technical quality-control of the optical density measured in each steps of the experiments:
- blank (BLANK),
- lysat (LYSATE),
- supernatant (SUPERNATANT1 and SUPERNATANT2).
This technical quality-control step checks:
- the variability among the duplicated optical density measures of each samples;
- the variability in the blank curves (intercept and slope estimates) among all experiments in a project.
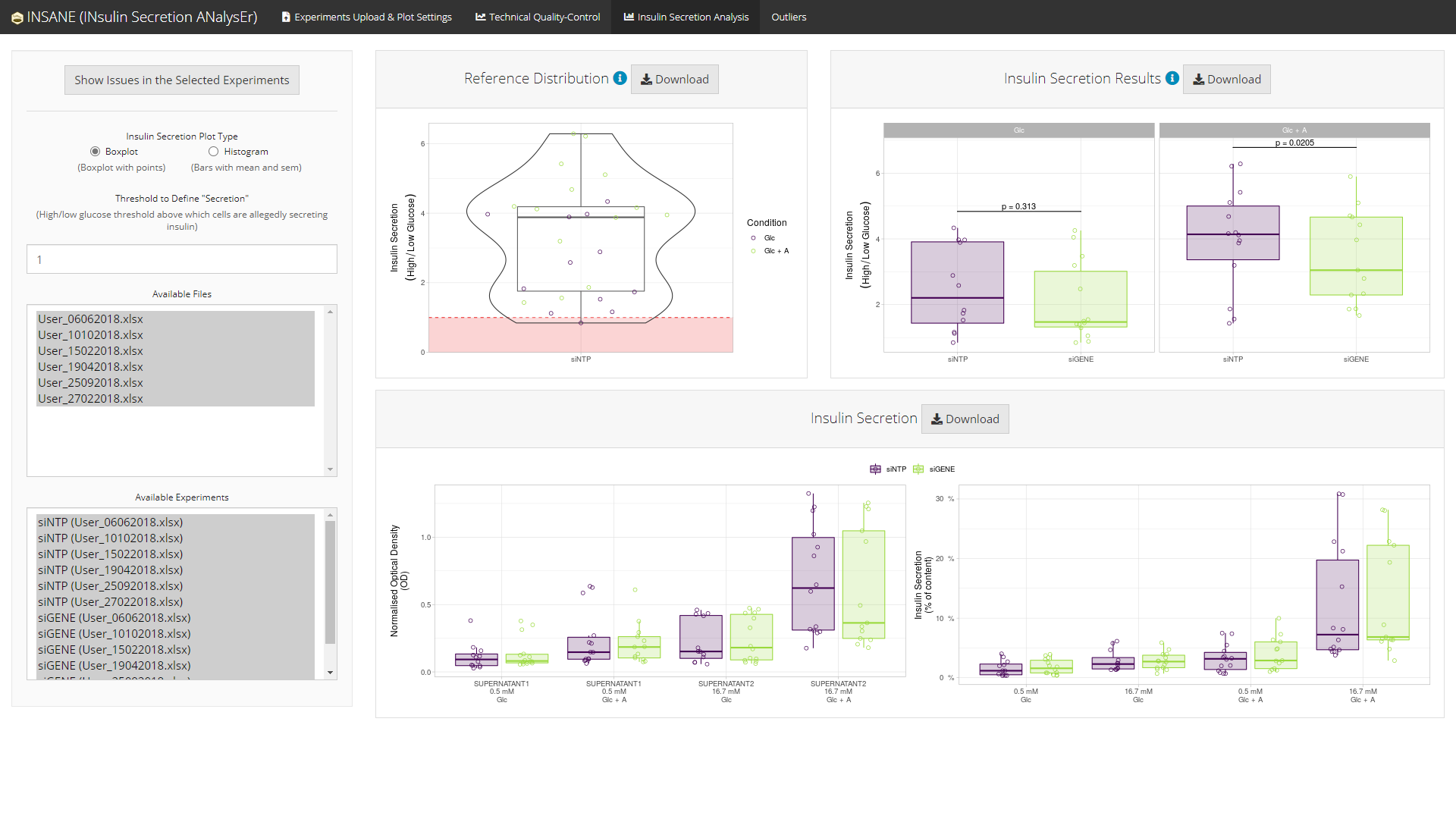
Statistical analyses (top)
insane performs statistical analyses of the experimental conditions, e.g., one silenced gene (siGENE) compared to an insulin secretion reference (siNTP) in two stimulation conditions (Glc and Glc + A).
Conditions are compared using a linear regression with Date and Operator as covariates (if needed) to control for heterogeneity.
Using all experiments in the selected project
Boxplot version
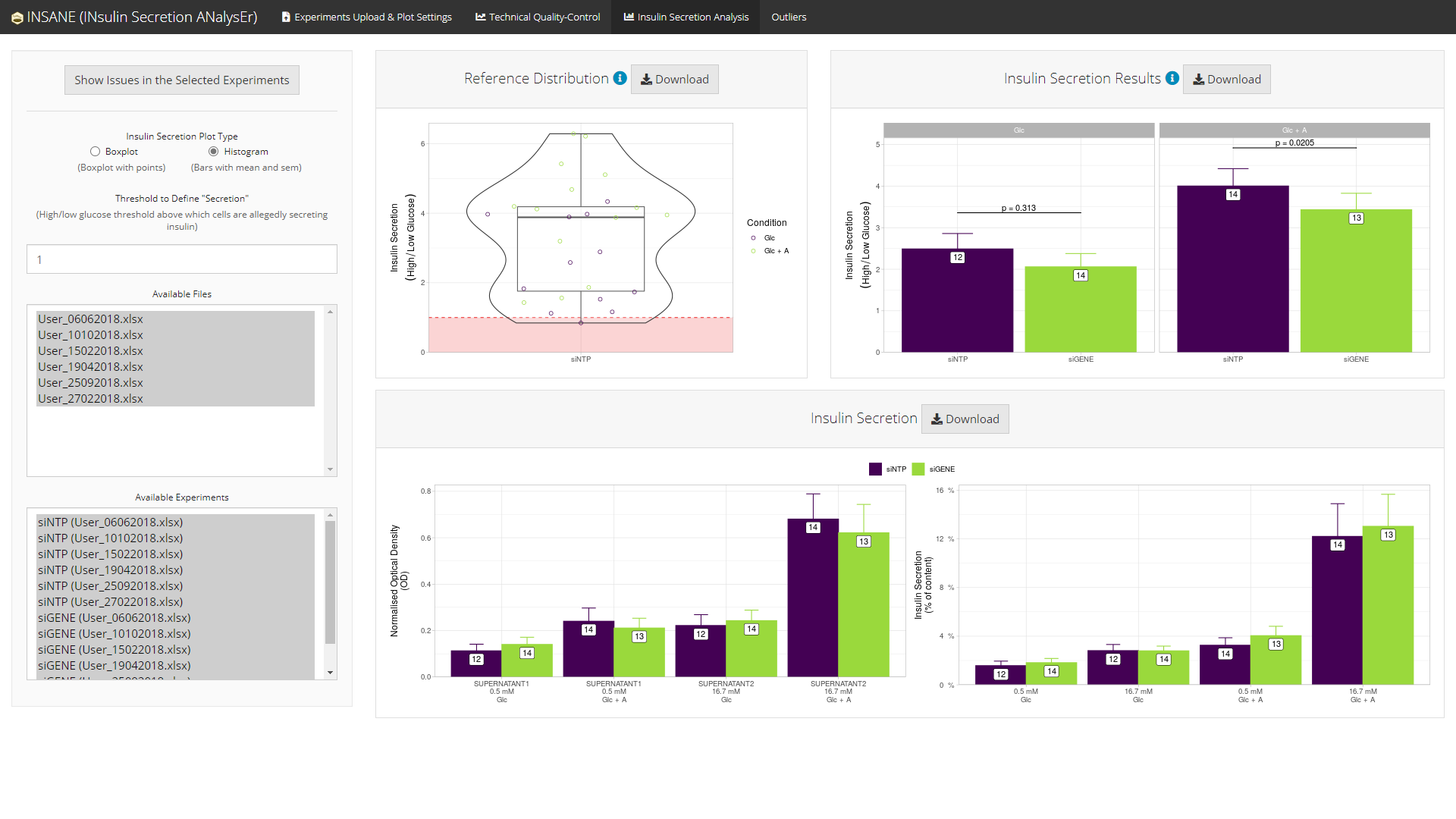
Histogram version
Using some of the experiments in the selected project
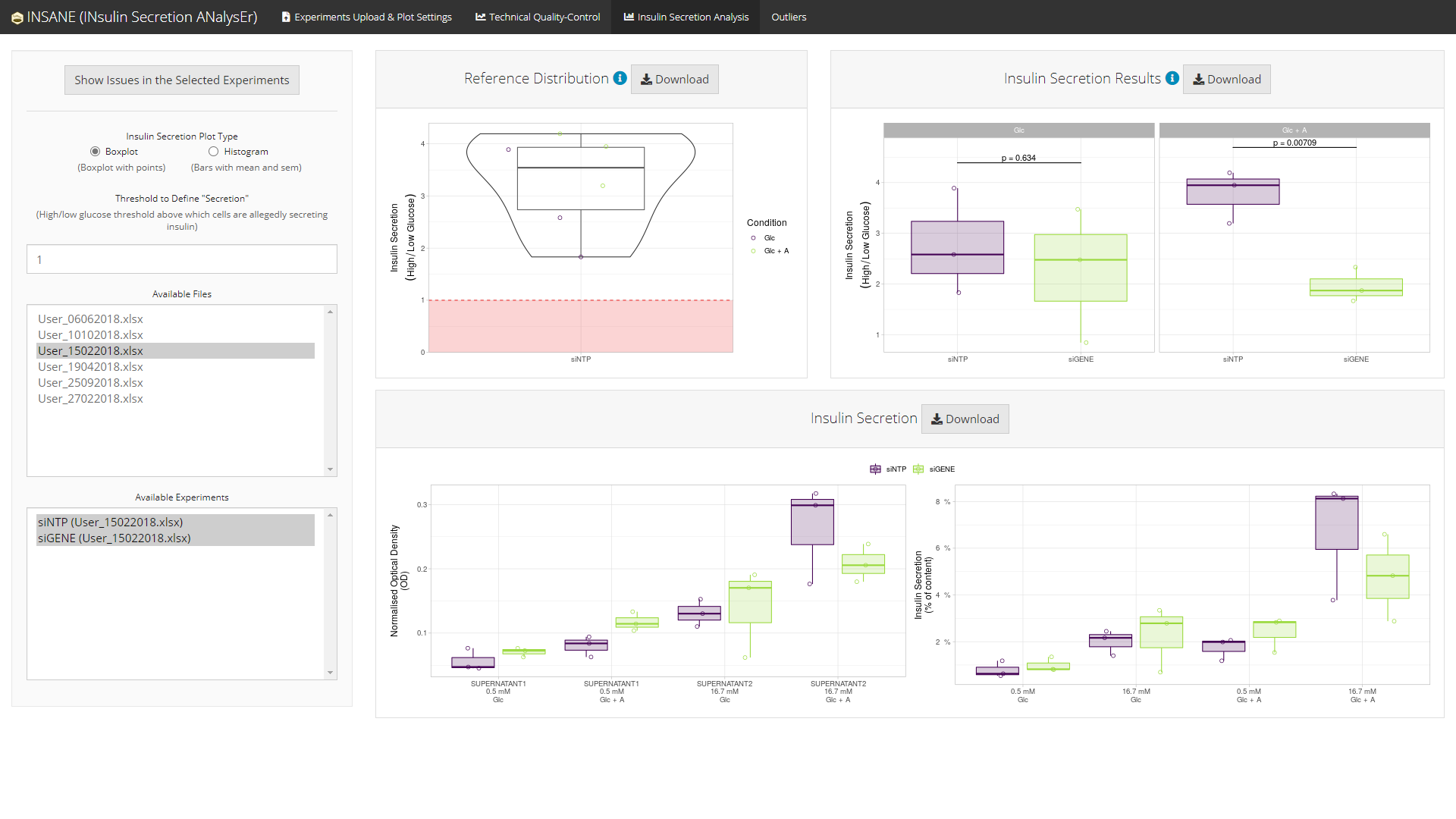
If and when some experiments are failing any of the technical quality-controls, a summary of the issues regarding the selected experiments can be displayed using the button Show Issues in the Selected Experiments.
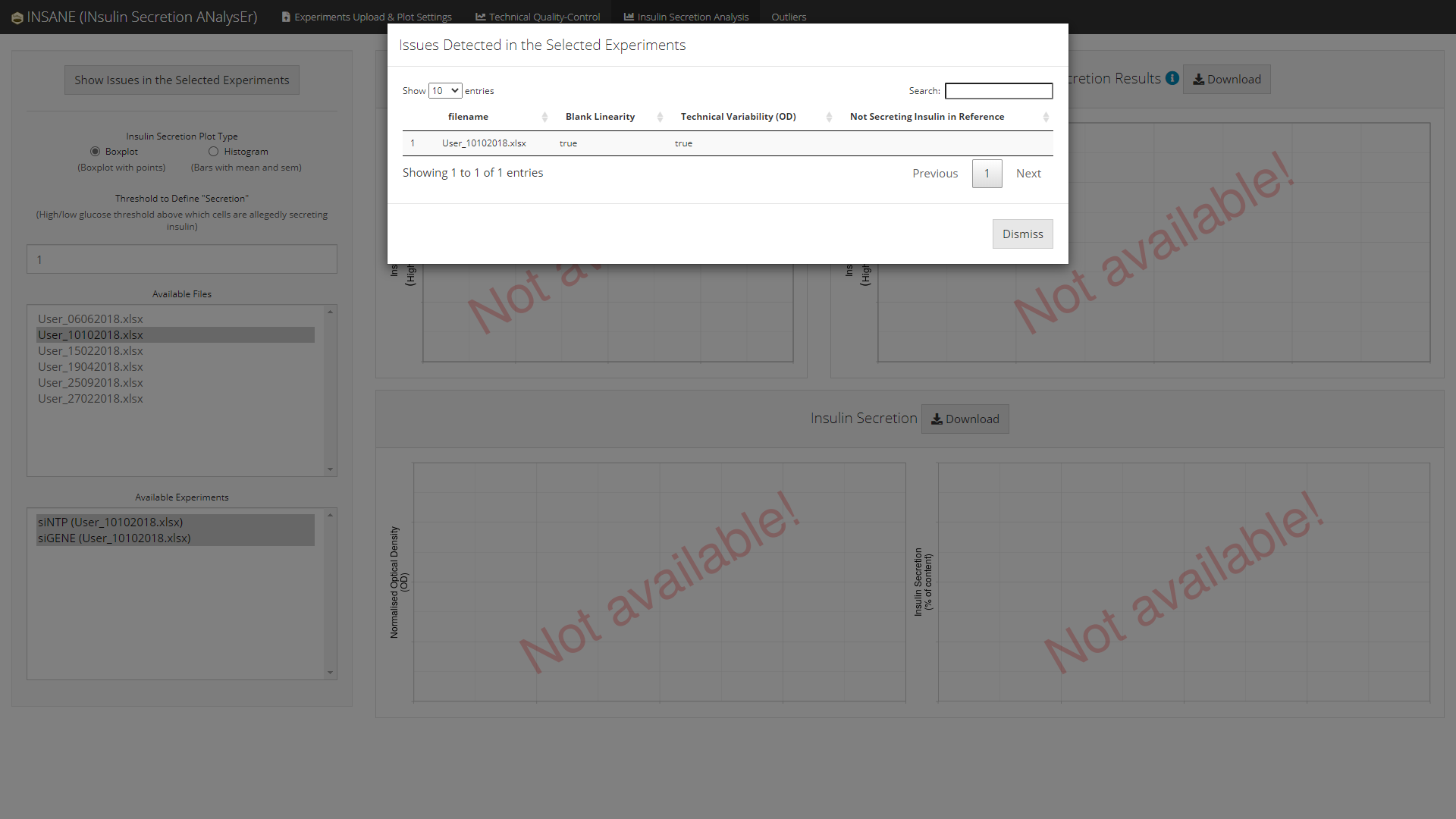
List of Outliers (Issues Detected) (top)
A comprehensive list of all issues detected in the selected project is available in an Outliers tab.
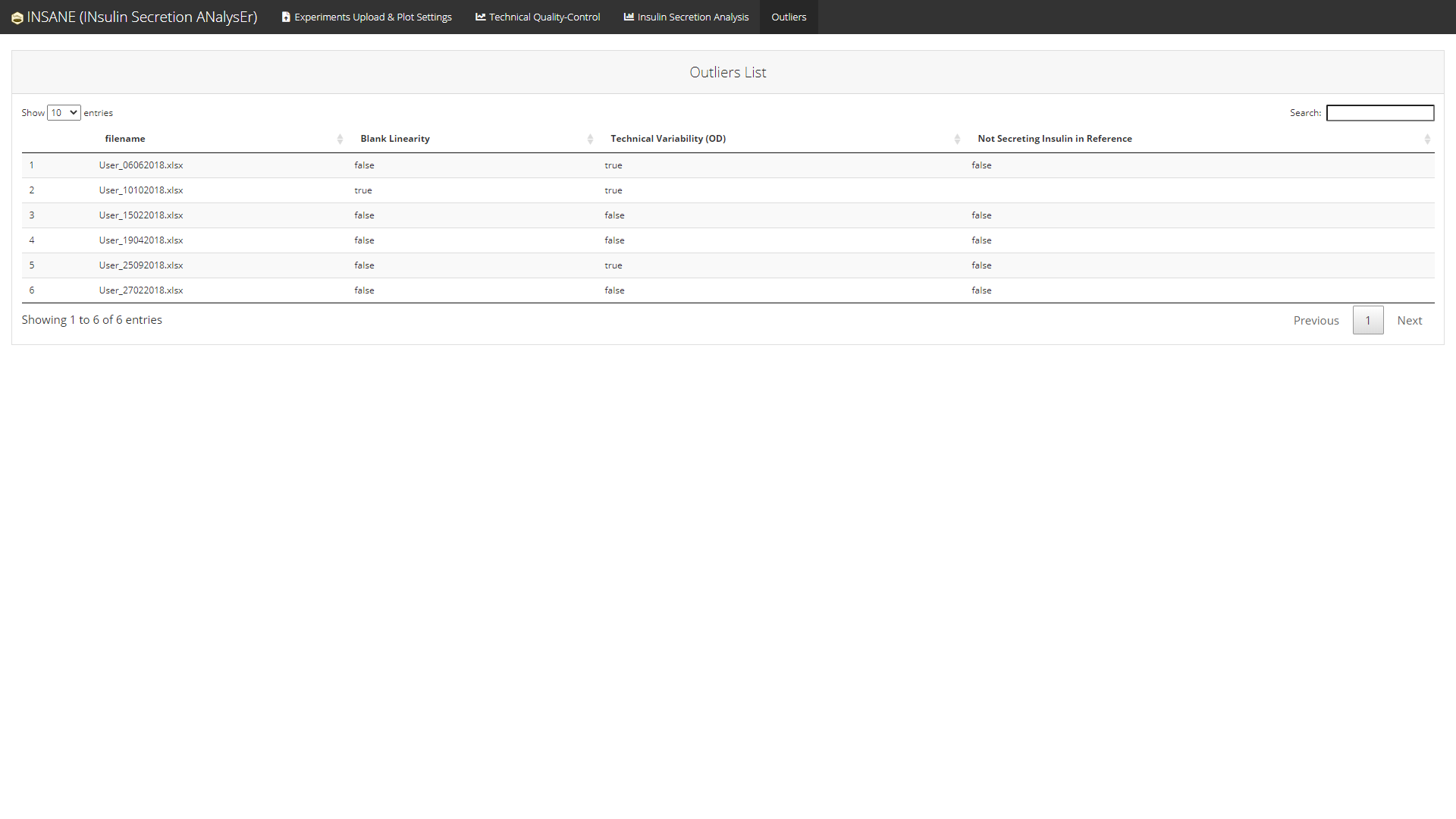
Note: The Outliers tab is displayed only if there is at least one issue in the selected project.
Getting help
If you encounter a clear bug, please file a minimal reproducible example on github.
For questions and other discussion, please contact the package maintainer.
Please note that this project is released with a Contributor Code of Conduct.
By participating in this project you agree to abide by its terms.
