Description
Tools to Easily Download Data from INSEE BDM Database.
Description
Using embedded sdmx queries, get the data of more than 150 000 insee series from 'bdm' macroeconomic database.
README.md
insee R package
Overview
The insee package contains tools to easily download data and metadata from INSEE main database (BDM).
Using embedded SDMX queries, get the data of more than 150 000 INSEE series.
Have a look at the detailed SDMX web service page on insee.fr.
This package is a contribution to reproducible research and public data transparency. It benefits from the developments made by INSEE's teams working on APIs.
Installation & Loading
# Get the development version from GitHub
# install.packages("devtools")
# devtools::install_github("pyr-opendatafr/R-Insee-Data")
# Get the CRAN version
install.packages("insee")
# examples below use tidyverse packages
library(tidyverse)
library(insee)
Examples & Tutorial
French GDP growth rate
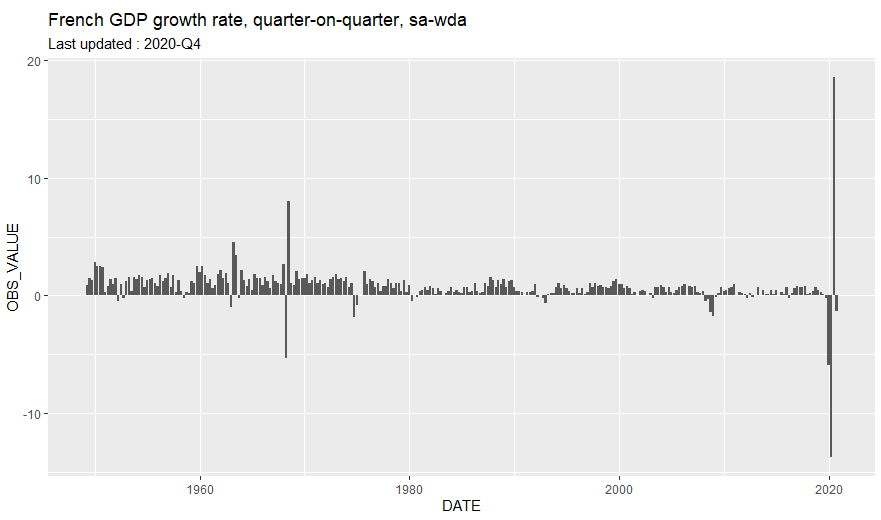
library(tidyverse)
library(insee)
dataset_list = get_dataset_list()
df_idbank_list_selected =
get_idbank_list("CNT-2014-PIB-EQB-RF") %>% # Gross domestic product balance
filter(FREQ == "T") %>% #quarter
add_insee_title() %>% #add titles
filter(OPERATION == "PIB") %>% #GDP
filter(NATURE == "TAUX") %>% #rate
filter(CORRECTION == "CVS-CJO") #SA-WDA, seasonally adjusted, working day adjusted
idbank = df_idbank_list_selected %>% pull(idbank)
data =
get_insee_idbank(idbank) %>%
add_insee_metadata()
ggplot(data, aes(x = DATE, y = OBS_VALUE)) +
geom_col() +
ggtitle("French GDP growth rate, quarter-on-quarter, sa-wda") +
labs(subtitle = sprintf("Last updated : %s", data$TIME_PERIOD[1]))
Deaths and Births
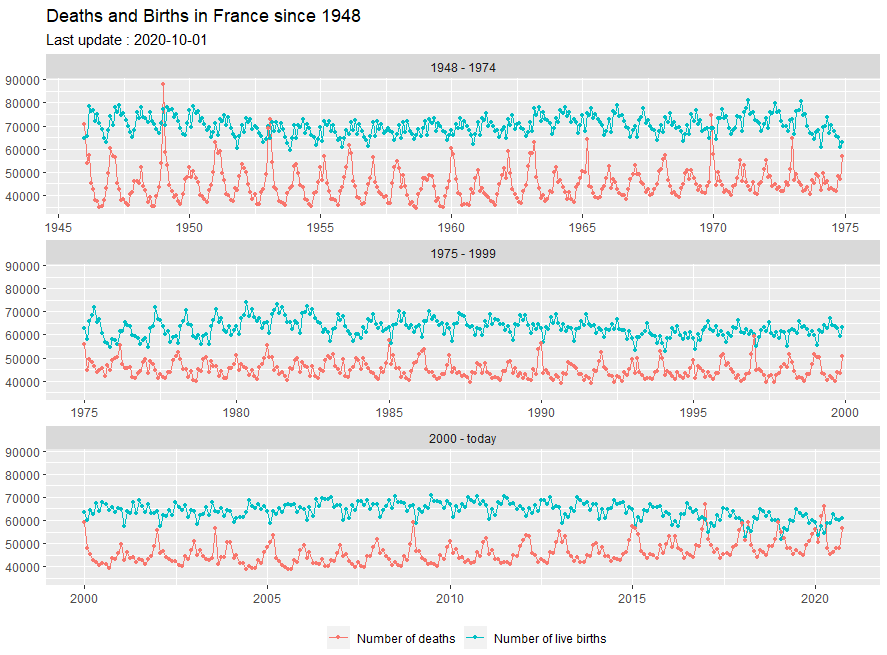
library(insee)
library(tidyverse)
insee_dataset = get_dataset_list()
list_idbank_selected =
get_idbank_list("DECES-MORTALITE", "NAISSANCES-FECONDITE") %>%
filter(FREQ == "M") %>% #monthly
filter(REF_AREA == "FM") %>% #metropolitan territory
filter(DEMOGRAPHIE %in% c("NAISS", "DECES"))
idbank_selected = list_idbank_selected %>% pull(idbank)
data =
get_insee_idbank(idbank_selected) %>%
split_title() %>%
mutate(period = case_when(DATE < "1975-01-01" ~ "1948 - 1974",
DATE >= "1975-01-01" & DATE < "2000-01-01" ~ "1975 - 1999",
DATE >= "2000-01-01" ~ "2000 - today"
))
x_dates = seq.Date(from = as.Date("1940-01-01"), to = Sys.Date(), by = "5 years")
last_date = data %>% pull(DATE) %>% max()
ggplot(data, aes(x = DATE, y = OBS_VALUE, colour = TITLE_EN2)) +
facet_wrap(~period, scales = "free_x", ncol = 1) +
geom_line() +
geom_point(size = 0.9) +
ggtitle("Deaths and Births in France since 1948") +
labs(subtitle = sprintf("Last update : %s", last_date)) +
scale_x_date(breaks = x_dates, date_labels = "%Y") +
theme(
legend.position = "bottom",
legend.title = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank()
)
Population Map
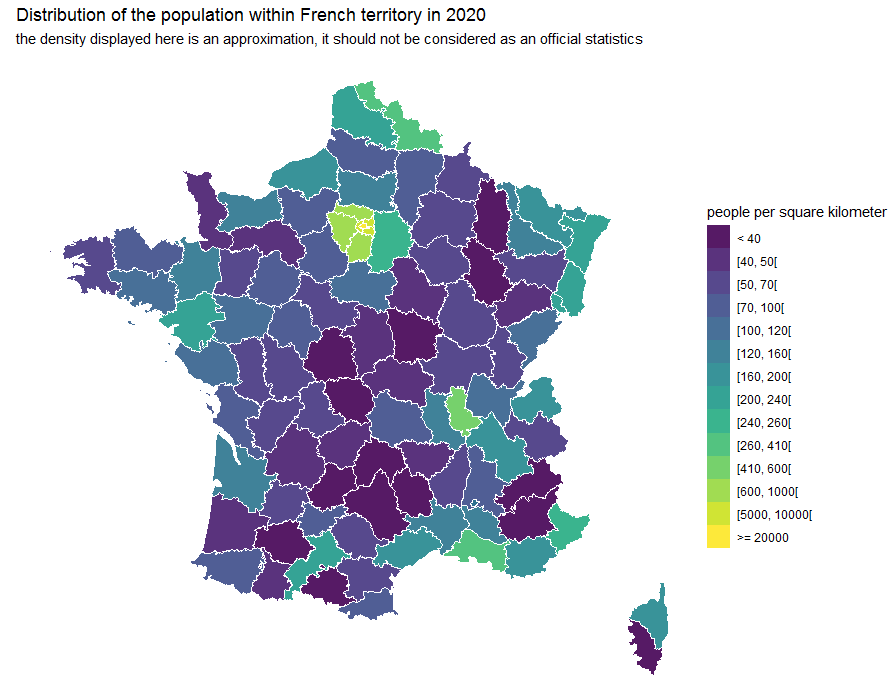
library(insee)
library(tidyverse)
library(raster)
library(rgdal)
library(geosphere)
library(broom)
library(viridis)
library(mapproj)
dataset_list = get_dataset_list()
list_idbank =
get_idbank_list("TCRED-ESTIMATIONS-POPULATION") %>%
filter(AGE == "00-") %>% #all ages
filter(SEXE == 0) %>% #men and women
filter(str_detect(REF_AREA, "^D")) %>% #select only departements
add_insee_title()
list_idbank_selected = list_idbank %>% pull(idbank)
# get population data by departement
pop = get_insee_idbank(list_idbank_selected)
#get departements' geographical limits
FranceMap <- raster::getData(name = "GADM", country = "FRA", level = 2)
# extract the population by departement in 2020
pop_plot = pop %>%
group_by(TITLE_EN) %>%
filter(DATE == "2020-01-01") %>%
mutate(dptm = gsub("D", "", REF_AREA)) %>%
filter(dptm %in% FranceMap@data$CC_2) %>%
mutate(dptm = factor(dptm, levels = FranceMap@data$CC_2)) %>%
arrange(dptm) %>%
mutate(id = dptm)
vec_pop = pop_plot %>% pull(OBS_VALUE)
# add population data to the departement object map
FranceMap@data$pop = vec_pop
get_area = function(long, lat){
area = areaPolygon(data.frame(long = long, lat = lat)) / 1000000
return(data.frame(area = area))
}
# extract the departements' limits from the spatial object and compute the surface
FranceMap_tidy_area <-
broom::tidy(FranceMap) %>%
group_by(id) %>%
group_modify(~get_area(long = .x$long, lat = .x$lat))
FranceMap_tidy <-
broom::tidy(FranceMap) %>%
left_join(FranceMap_tidy_area)
# mapping table
dptm_df = data.frame(dptm = FranceMap@data$CC_2,
dptm_name = FranceMap@data$NAME_2,
pop = FranceMap@data$pop,
id = rownames(FranceMap@data))
FranceMap_tidy_final_all =
FranceMap_tidy %>%
left_join(dptm_df, by = "id") %>%
mutate(pop_density = pop/area) %>%
mutate(density_range = case_when(pop_density < 40 ~ "< 40",
pop_density >= 40 & pop_density < 50 ~ "[40, 50]",
pop_density >= 50 & pop_density < 70 ~ "[50, 70]",
pop_density >= 70 & pop_density < 100 ~ "[70, 100]",
pop_density >= 100 & pop_density < 120 ~ "[100, 120]",
pop_density >= 120 & pop_density < 160 ~ "[120, 160]",
pop_density >= 160 & pop_density < 200 ~ "[160, 200]",
pop_density >= 200 & pop_density < 240 ~ "[200, 240]",
pop_density >= 240 & pop_density < 260 ~ "[240, 260]",
pop_density >= 260 & pop_density < 410 ~ "[260, 410]",
pop_density >= 410 & pop_density < 600 ~ "[410, 600]",
pop_density >= 600 & pop_density < 1000 ~ "[600, 1000]",
pop_density >= 1000 & pop_density < 5000 ~ "[1000, 5000]",
pop_density >= 5000 & pop_density < 10000 ~ "[5000, 10000]",
pop_density >= 20000 ~ ">= 20000"
)) %>%
mutate(`people per square kilometer` = factor(density_range,
levels = c("< 40","[40, 50]", "[50, 70]","[70, 100]",
"[100, 120]", "[120, 160]", "[160, 200]",
"[200, 240]", "[240, 260]", "[260, 410]",
"[410, 600]", "[600, 1000]", "[1000, 5000]",
"[5000, 10000]", ">= 20000")))
ggplot(data = FranceMap_tidy_final_all,
aes(fill = `people per square kilometer`, x = long, y = lat, group = group) ,
size = 0, alpha = 0.9) +
geom_polygon() +
geom_path(colour = "white") +
coord_map() +
theme_void() +
scale_fill_viridis(discrete = T) +
ggtitle("Distribution of the population within French territory in 2020") +
labs(subtitle = "the density displayed here is an approximation, it should not be considered as an official statistics")
How to avoid proxy issues ?
Sys.setenv(http_proxy = "my_proxy_server")
Sys.setenv(https_proxy = "my_proxy_server")
Support
Feel free to open an issue with any question about this package using https://github.com/pyr-opendatafr/R-Insee-Data Github repository.
