Isotope Origin Clustering and Assignment Tools.
isocat
isocat: Isotope Clustering and Assignment Tools
The isocat package provides multiple tools in R for creating and quantitatively analyzing and summarizing probability-of-origin surfaces generated from stable isotope analyses of animal tissue. The package includes functions to create probability-of-origin surfaces, quantitatively compare and summarize the origins of individuals, and create and validate post-processing surfaces useful for interpreting individual origins.
Many of isocat's functionalities are explored in Campbell et al. (2020) (https://doi.org/10.1515/ami-2020-0004)
Overview
isocat relates the stable isotope ratios of animal tissues to environmental isotope data to create probability-of-origin maps for individuals.
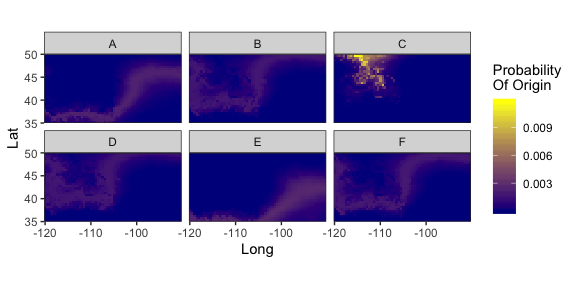
Probability-of-origin maps can be quantitatively compared, and individual origins compared and clustered.
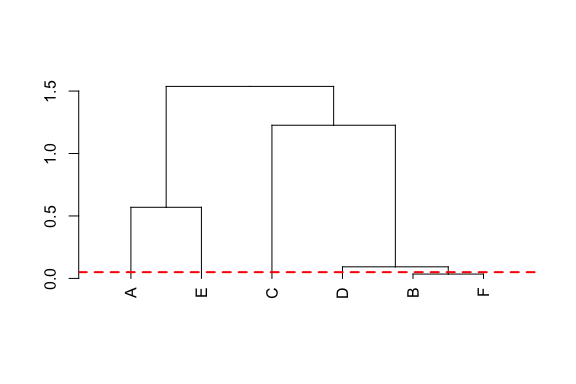
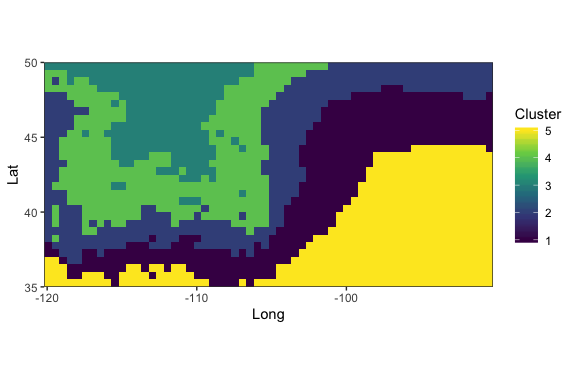
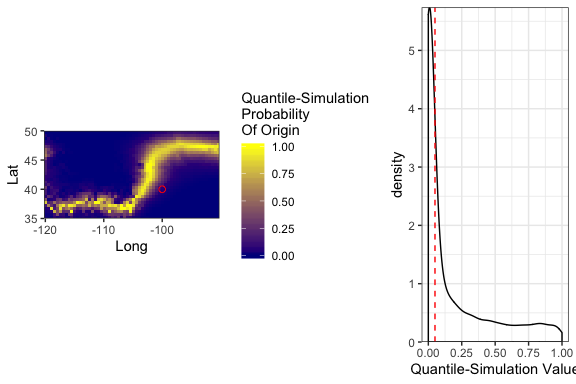
Probability-of-origin surfaces can also be transformed to several common post-processing surfaces to facilitate interpretation of individual origins, including cumulative-sum, odds-ratio, probability quantile, and quantile-simulation surfaces. Functions are also available to find untransformed and post-processing probability-of-origin values at specific sampling sites.
For more information, and for reproducible examples, please see isocat's vignette.
Installation
CRAN
isocat is available on CRAN. Install using install.packages("isocat")
GitHub
The development version of isocat is available on GitHub. To install without a vignette, use: devtools::install_github("cjcampbell/isocat")
The isocat package provides multiple tools in R for creating and quantitatively analyzing and summarizing probability-of-origin surfaces generated from stable isotope analyses of animal tissue.
Vignette
To install isocat and its vignette directly from github, use the following code in R when using devtools >= 2.0.0:
devtools::install_github("cjcampbell/isocat", build = TRUE, force = TRUE, build_opts = c("--no-resave-data", "--no-manual"))
The vignette requires the certain packages to be installed in order to compile. The code chunk below will automatically check for and install packages needed to form the vignette.
vignettePackages <- c("kableExtra", "pvclust", "rasterVis", "ggplot2", "viridisLite", "gridExtra", "dplyr", "dendextend", "isocat")
lapply(vignettePackages, function(pckg) {
if (!require(pckg, character.only = TRUE, quietly = FALSE)) {
install.packages(x, dependencies = TRUE)
require(pckg)
}
})
}