Junction Tree Inference.
jti: Junction Tree Inference 
About
The jti package (pronounced ‘yeti’) is a memory efficient implementaion of the junction tree algorithm (JTA) using the Lauritzen-Spiegelhalter scheme. Why is it memory efficient? Because we use a for the potentials which enable us to handle large and complex graphs where the variables can have an arbitrary large number of levels. The jti package is a big part of the software paper
.
Installation
Current stable release from CRAN:
install.packages("jti")
Development version (see README.md for new features):
devtools::install_github("mlindsk/jti", build_vignettes = FALSE)
Libraries
library(jti)
library(igraph)
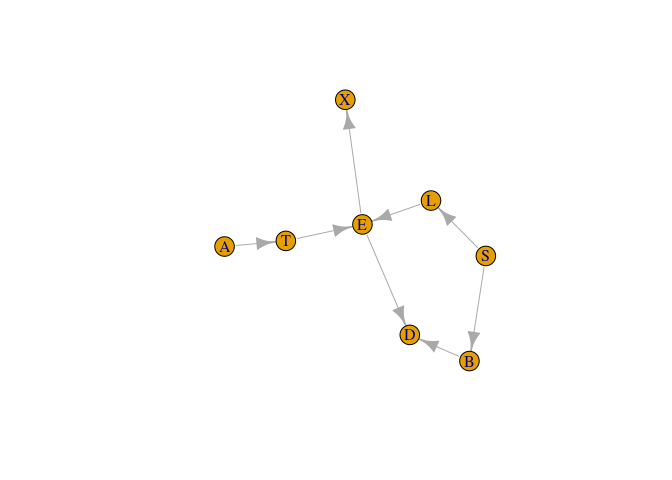
Setting up the network
el <- matrix(c(
"A", "T",
"T", "E",
"S", "L",
"S", "B",
"L", "E",
"E", "X",
"E", "D",
"B", "D"),
nc = 2,
byrow = TRUE
)
g <- igraph::graph_from_edgelist(el)
plot(g)
We use the asia data; see the man page (?asia)
Compilation
Checking and conversion
cl <- cpt_list(asia, g)
cl
#> List of CPTs
#> -------------------------
#> P( A )
#> P( T | A )
#> P( E | T, L )
#> P( S )
#> P( L | S )
#> P( B | S )
#> P( X | E )
#> P( D | E, B )
#>
#> <cpt_list, list>
#> -------------------------
Compilation
cp <- compile(cl)
cp
#> Compiled network
#> -------------------------
#> Nodes: 8
#> Cliques: 6
#> - max: 3
#> - min: 2
#> - avg: 2.67
#> <charge, list>
#> -------------------------
# plot(get_graph(cp)) # Should give the same as plot(g)
After the network has been compiled, the graph has been triangulated and moralized. Furthermore, all conditional probability tables (CPTs) has been designated to one of the cliques (in the triangulated and moralized graph).
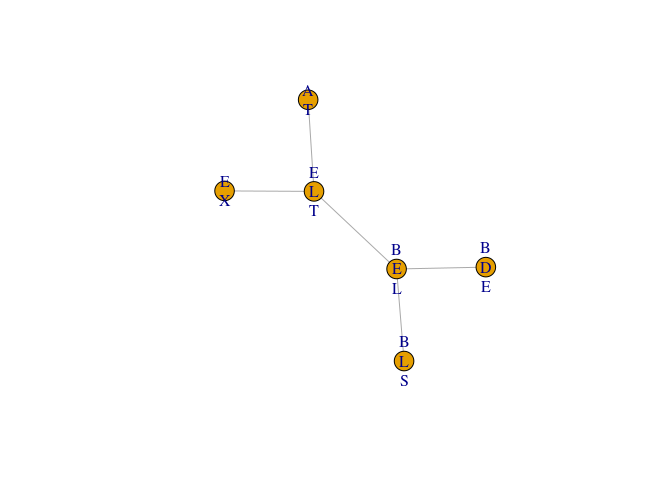
Example 1: sum-flow without evidence
jt1 <- jt(cp)
jt1
#> Junction Tree
#> -------------------------
#> Propagated: full
#> Flow: sum
#> Cliques: 6
#> - max: 3
#> - min: 2
#> - avg: 2.67
#> <jt, list>
#> -------------------------
plot(jt1)
 Query probabilities
Query probabilities query_belief(jt1, c("E", "L", "T"))
#> $E
#> E
#> n y
#> 0.9257808 0.0742192
#>
#> $L
#> L
#> n y
#> 0.934 0.066
#>
#> $T
#> T
#> n y
#> 0.9912 0.0088
query_belief(jt1, c("B", "D", "E"), type = "joint")
#> , , B = y
#>
#> E
#> D n y
#> y 0.36261346 0.041523361
#> n 0.09856873 0.007094444
#>
#> , , B = n
#>
#> E
#> D n y
#> y 0.04637955 0.018500278
#> n 0.41821906 0.007101117
It should be noticed, that the above could also have been achieved by
jt1 <- jt(cp, propagate = "no")
jt1 <- propagate(jt1, prop = "full")
That is; it is possible to postpone the actual propagation.
Example 2: sum-flow with evidence
e2 <- c(A = "y", X = "n")
jt2 <- jt(cp, e2)
query_belief(jt2, c("B", "D", "E"), type = "joint")
#> , , B = y
#>
#> E
#> D n y
#> y 0.3914092 3.615182e-04
#> n 0.1063963 6.176693e-05
#>
#> , , B = n
#>
#> E
#> D n y
#> y 0.05006263 2.009085e-04
#> n 0.45143057 7.711638e-05
Notice that, the configuration (D,E,B) = (y,y,n) has changed dramatically as a consequence of the evidence. We can get the probability of the evidence:
query_evidence(jt2)
#> [1] 0.007152638
Example 3: max-flow without evidence
jt3 <- jt(cp, flow = "max")
mpe(jt3)
#> A T E L S B X D
#> "n" "n" "n" "n" "n" "n" "n" "n"
Example 4: max-flow with evidence
e4 <- c(T = "y", X = "y", D = "y")
jt4 <- jt(cp, e4, flow = "max")
mpe(jt4)
#> A T E L S B X D
#> "n" "y" "y" "n" "y" "y" "y" "y"
Notice, that T, E, S, B, X and D has changed from "n" to "y" as a consequence of the new evidence e4.
Example 5: specifying a root node and only collect to save run time
cp5 <- compile(cpt_list(asia, g) , root_node = "X")
jt5 <- jt(cp5, propagate = "collect")
We can only query from the variables in the root clique now but we have ensured that the node of interest, “X”, does indeed live in this clique. The variables are found using get_clique_root.
query_belief(jt5, get_clique_root(jt5), "joint")
#> E
#> X n y
#> n 0.88559032 0.0004011849
#> y 0.04019048 0.0738180151
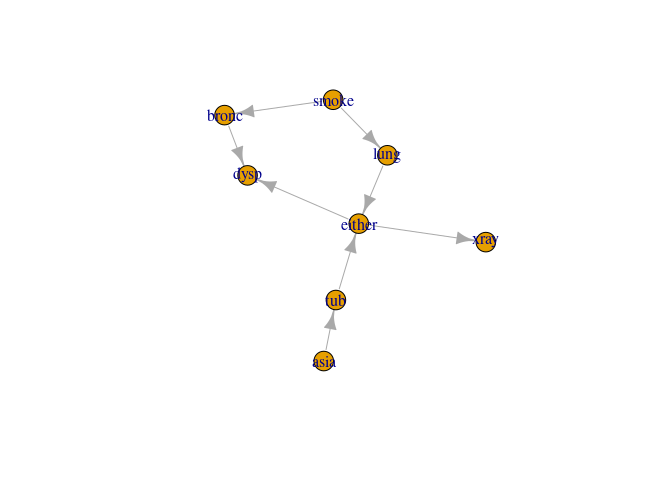
Example 6: Compiling from a list of conditional probabilities
- We need a list with CPTs which we extract from the asia2 object
- the list must be named with child nodes
- The elements need to be array-like objects
cl <- cpt_list(asia2)
cp6 <- compile(cl)
Inspection; see if the graph correspond to the cpts
plot(get_graph(cp6))
This time we specify that no propagation should be performed
jt6 <- jt(cp6, propagate = "no")
We can now inspect the collecting junction tree and see which cliques are leaves and parents
plot(jt6)
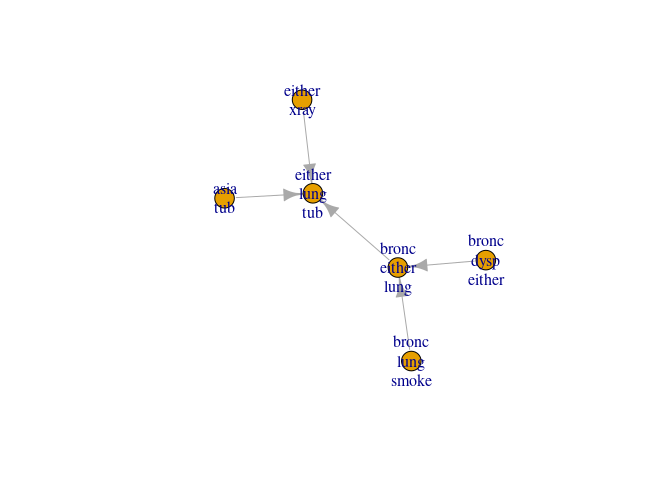
get_cliques(jt6)
#> $C1
#> [1] "asia" "tub"
#>
#> $C2
#> [1] "either" "lung" "tub"
#>
#> $C3
#> [1] "bronc" "either" "lung"
#>
#> $C4
#> [1] "bronc" "lung" "smoke"
#>
#> $C5
#> [1] "bronc" "dysp" "either"
#>
#> $C6
#> [1] "either" "xray"
get_clique_root(jt6)
#> [1] "either" "lung" "tub"
leaves(jt6)
#> [1] 1 4 5 6
unlist(parents(jt6))
#> [1] 2 3 3 2
That is; - clique 2 is parent of clique 1 - clique 3 is parent of clique 4 etc.
Next, we send the messages from the leaves to the parents
jt6 <- send_messages(jt6)
Inspect again
plot(jt6)
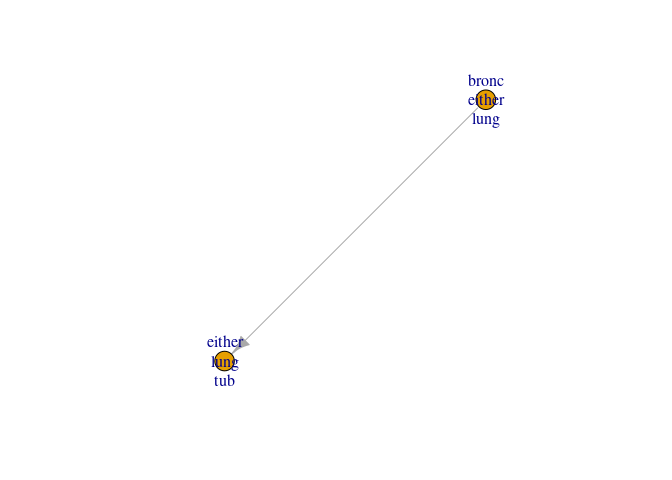
Send the last message to the root and inspect
jt6 <- send_messages(jt6)
plot(jt6)
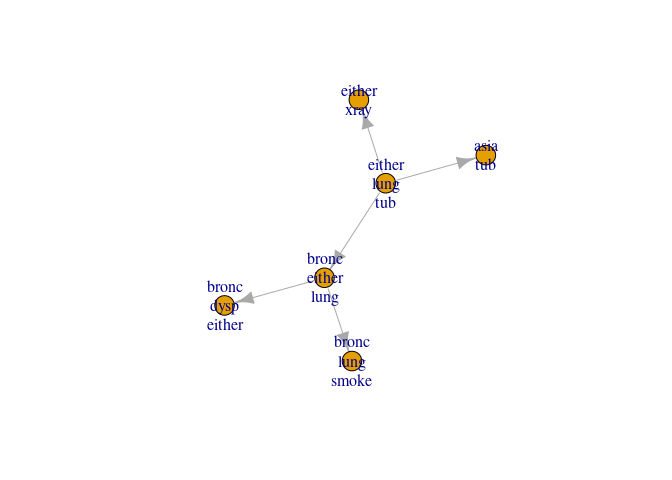
The arrows are now reversed and the outwards (distribute) phase begins
leaves(jt6)
#> [1] 2
parents(jt6)
#> [[1]]
#> [1] 1 3 6
Clique 2 (the root) is now a leave and it has 1, 3 and 6 as parents. Finishing the message passing
jt6 <- send_messages(jt6)
jt6 <- send_messages(jt6)
Queries can now be performed as normal
query_belief(jt6, c("either", "tub"), "joint")
#> tub
#> either yes no
#> yes 0.0104 0.054428
#> no 0.0000 0.935172
Example 7: Fitting a decomposable model and apply JTA
We use the ess package (on CRAN), found at https://github.com/mlindsk/ess, to fit an undirected decomposable graph to data.
library(ess)
g7 <- ess::fit_graph(asia, trace = FALSE)
ig7 <- ess::as_igraph(g7)
cp7 <- compile(pot_list(asia, ig7))
jt7 <- jt(cp7)
query_belief(jt7, get_cliques(jt7)[[4]], type = "joint")
#> , , T = n
#>
#> L
#> E n y
#> n 0.999967 0.000000e+00
#> y 0.000000 2.930828e-05
#>
#> , , T = y
#>
#> L
#> E n y
#> n 0.000000e+00 0.000000e+00
#> y 3.657762e-06 6.286238e-09