Langevin Tour.
langevitour
langevitour is an HTML widget that randomly tours projections of a high-dimensional dataset with an animated scatter-plot. The user can manipulate the plot to use specified axes, or turn on Guided Tour mode to perform projection pursuit, finding an informative projection of the data. Groups within the data can be hidden or shown, as can particular axes. Known projections of interest can be added as "extra axes" and also manipulated. The widget can be used from within R or Python, or included in a self-contained Rmarkdown document, or a Shiny app, or used directly from Javascript.
langevitour is a twist on the "tour" concept from software such as XGobi, GGobi, tourr, ferrn, liminal, detourr, spinifex, and loon.tour. The new element in langevitour is the use of Langevin Dynamics to generate the sequence of projections.
langevitour is described in:
Harrison, Paul. 2023. "langevitour: Smooth Interactive Touring of High Dimensions, Demonstrated with scRNA-Seq Data." The R Journal 15 (2): 206–219. https://doi.org/10.32614/RJ-2023-046.
Further material:
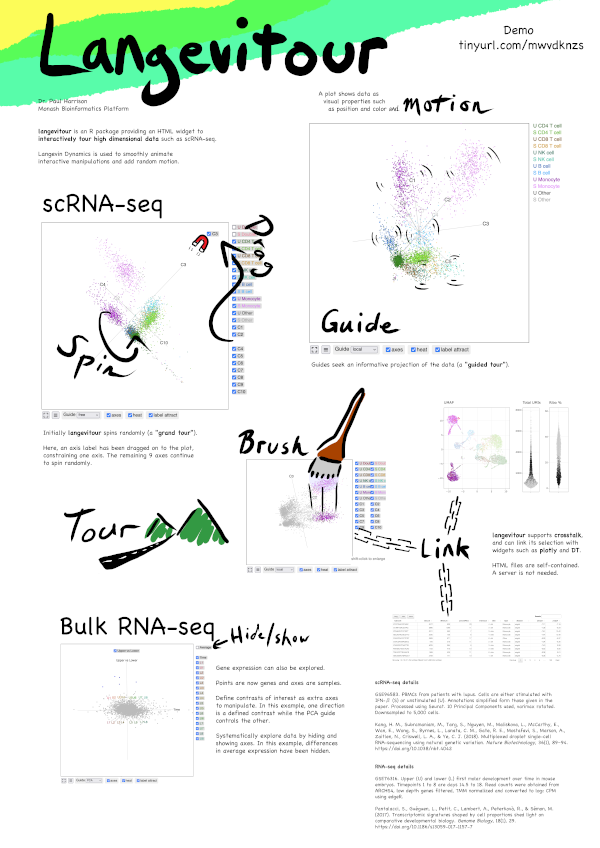
ABACBS Conference 2022 poster (large image) and demo
ABACBS Seminar 2022 slides
useR! 2022 conference slides and video (extended edition)
IASC-ARS 2023 slides and short video for Visualising high-dimensional genomics data: what Non-Linear Dimension Reduction hides and misrepresents. Demonstrates some advanced tricks setting the state of the widget using buttons in a Quarto presentation.
R installation
Release version:
install.packages("langevitour")
Development version:
remotes::install_github("pfh/langevitour")
To build the documentation site:
install.packages(c("devtools", "pkgdown", "BiocManager"))
devtools::install_dev_deps()
BiocManager::install(c("airway", "org.Hs.eg.db", "edgeR", "limma"))
pkgdown::build_site()
R usage
Example:
library(langevitour)
data(zeiselPC)
langevitour(zeiselPC[,-1], zeiselPC$type)
JavaScript usage
- Get started by viewing source on this example.
- JavaScript documentation.
The minified and bundled version can be found in inst/htmlwidgets/lib/langevitour-pack.js.
ESM module with npm
If using node and npm for development, langevitour can be added with:
npm install langevitour
This provides the widget as a modern ESM module. In your HTML page you can import it with:
<script type="module">
import { Langevitour } from "langevitour";
// ...
</script>
You'll need to use a packager such as parcel or webpack to use this. Please tell me if you run into any problems, I am fairly new to Javascript development.
ESM module without npm
To avoid using npm, you could use skypack.dev. You will still need to serve your page with some sort of web-server, such as python3 -m http.server.
<script type="module">
import { Langevitour } from "https://cdn.skypack.dev/langevitour";
// ...
</script>
Here is an example using skypack in an Observable Notebook.
JavaScript development
langevitour is written in TypeScript, which is compiled to JavaScipt, and then Webpack is used to produce a minified and bundled version. To make changes to the JavaScript side of langevitour, you will need to install node, which includes the npm package manager. npm can then install the necessary build tools and dependencies. Build scripts are defined in package.json and used as below.
git clone https://github.com/pfh/langevitour.git
cd langevitour
# Install required packages
npm install
# ... edit source in src/ directory ...
# Compile TypeScript modules in src/ to JavaScript modules in lib/.
# Produce minified bundle inst/htmlwidgets/lib/langevitour.js
npm run js-build
# Complete Javascript+R build and documentation process.
npm run build
For example, to define a new guide you would:
- Add a new gradient function in
ts/guides.ts. - Add it to the
gradTableints/guides.ts. - Add it to the
guideSelectselect box ints/langevitour.ts. - Run
npm run js-buildand the new guide should appear when you loadexample.html.
Python installation
pip install langevitour
Python usage
import numpy as np
from langevitour import Langevitour
# Generate a sample dataset
X = []
group = []
n = 20000
def r():
return np.random.normal(0, 0.02)
for i in range(n):
a = i/n * np.pi * 2
X.append([
10 + np.sin(a)/3 + r(),
20 + np.sin(a*2)/3 + r(),
30 + np.sin(a*3)/3,
40 + np.sin(a*4)/3,
50 + np.sin(a*5)/3
])
group.append(int(i*4/n))
# Extra axes (specified as columns of a matrix)
extra_axes = [[1], [2], [0], [0], [0]]
extra_axes_names = ["V1+2*V2"]
tour = Langevitour(
X,
group=group,
extra_axes=extra_axes,
extra_axes_names=extra_axes_names,
point_size=1,
)
tour.write_html("langevitour_plot.html")
langevitour also works in jupyter notebooks.
Authors
The Javascript and R package are written by Paul Harrison. The Python package was kindly contributed by Wytamma Wirth.
Copyright
Langevitour is free software made available under the MIT license. Included libraries jStat and SVD-JS are also provided under the MIT license. Included library D3 is provided under the ISC license.