Lemna Ecotox Effect Model.
lemna
Overview
lemna is a software package for the language R. It implements model equations and default parameters to simulate the toxicokinetic-toxicodynamic (TKTD) model of the Lemna aquatic plant. Lemna is a standard test macrophyte used in ecotox effect studies. The model was described and published by the SETAC Europe Interest Group Effect Modeling (Klein et al. 2022). It is a refined description of the Lemna TKTD model published by Schmitt et al. (2013). This package contains the model’s reference implementation which is provided by the SETAC interest group.
Installation
## install directly from CRAN
install.packages("lemna")
## install latest development version from GitHub
#install.packages("remotes")
remotes::install_github("nkehrein/lemna", dependencies=TRUE)
Usage
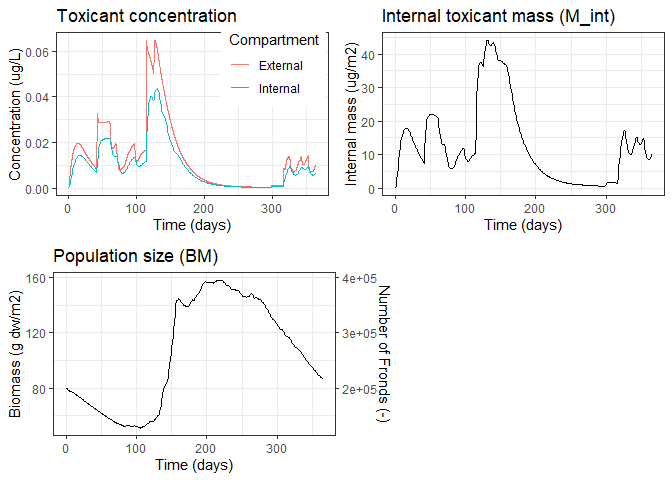
The lemna package provides model equations and some useful helpers to simulate the growth of Lemna (duckweed) aquatic plant populations. A simulation is run by using the lemna() function. The required scenario data are either supplied individually on function call or are passed as a pre-defined scenario object, such as the focusd1 sample scenario:
library(lemna)
# simulate sample scenario
result <- lemna(focusd1)
head(result)
#> time BM M_int C_int FrondNo
#> 1 0 80.00000 0.0000000 0.0000000000 200000.0
#> 2 1 79.48177 0.6614503 0.0004983256 198704.4
#> 3 2 79.00414 2.2061048 0.0016720907 197510.3
#> 4 3 78.57538 4.1750616 0.0031817050 196438.5
#> 5 4 78.23362 6.2899397 0.0048143381 195584.0
#> 6 5 78.01035 8.3810150 0.0064332127 195025.9
plot(result)
Learning lemna
The package contains two vignettes that may help you getting started:
- Introduction to the Lemna package
A general Tutorial and guide to the package functions - Lemna model verification
A verification of the model implementation against results of the Schmitt et al. implementation. Contains advanced workflows of package features.
License
The package and its source code is free and open-source software available under the MIT license.
Issues
If you find any issues or bugs within the package, please create a new issue on GitHub.
References
- Klein J., Cedergreen N., Heine S., Kehrein N., Reichenberger S., Rendal C., Schmitt W., Hommen U., 2022: Refined description of the Lemna TKTD growth model based on Schmitt et al. (2013) – equation system and default parameters, implementation in R. Report of the working group Lemna of the SETAC Europe Interest Group Effect Modeling. Version 1.1, uploaded on 09 May 2022. https://www.setac.org/group/SEIGEffectModeling
- Schmitt W., Bruns E., Dollinger M., Sowig P., 2013: Mechanistic TK/TD-model simulating the effect of growth inhibitors on Lemna populations. Ecol Model 255, pp. 1-10. DOI: 10.1016/j.ecolmodel.2013.01.017