Description
Real-Time PCR Data Sets by Lievens et al. (2012).
Description
Real-time quantitative polymerase chain reaction (qPCR) data sets by Lievens et al. (2012) <doi:10.1093/nar/gkr775>. Provides one single tabular tidy data set in long format, encompassing three dilution series, targeted against the soybean Lectin endogene. Each dilution series was assayed in one of the following PCR-efficiency-modifying conditions: no PCR inhibition, inhibition by isopropanol and inhibition by tannic acid. The inhibitors were co-diluted along with the dilution series. The co-dilution series consists of a five-point, five-fold serial dilution. For each concentration there are 18 replicates. Each amplification curve is 60 cycles long. Original raw data file is available at the Supplementary Data section at Nucleic Acids Research Online <doi:10.1093/nar/gkr775>.
README.md
lievens
{lievens} provides Real-Time PCR Data Sets by Lievens et al. (2012).
Installation
install.packages("lievens")
Data
Each sample group is defined by the inhibitor (“none”, “isopropanol”, “tannic acid”), respective inhibitor concentration, and initial amplicon copy number. Note that inhibitor concentration and initial copy number of the amplicon are not independent of each other as concentrations are obtained by co-dilution.
library(lievens)
library(ggplot2)
library(dplyr, warn.conflicts = FALSE)
lievens |>
dplyr::distinct(plate, inhibitor, inhibitor_conc, copies, dilution)
#> # A tibble: 15 × 5
#> plate inhibitor inhibitor_conc copies dilution
#> <fct> <fct> <dbl> <int> <int>
#> 1 soy none 0 100000 1
#> 2 soy none 0 20000 5
#> 3 soy none 0 4000 25
#> 4 soy none 0 800 125
#> 5 soy none 0 160 625
#> 6 soy+isopropanol isopropanol 2.5 100000 1
#> 7 soy+isopropanol isopropanol 0.5 20000 5
#> 8 soy+isopropanol isopropanol 0.1 4000 25
#> 9 soy+isopropanol isopropanol 0.02 800 125
#> 10 soy+isopropanol isopropanol 0.004 160 625
#> 11 soy+tannic acid tannic acid 0.2 100000 1
#> 12 soy+tannic acid tannic acid 0.04 20000 5
#> 13 soy+tannic acid tannic acid 0.008 4000 25
#> 14 soy+tannic acid tannic acid 0.0016 800 125
#> 15 soy+tannic acid tannic acid 0.0032 160 625
Here is the number of replicates per group:
lievens |>
dplyr::distinct(plate, inhibitor, inhibitor_conc, copies, dilution, replicate) |>
dplyr::count(plate, inhibitor, inhibitor_conc, copies, dilution) |>
print(n = Inf)
#> # A tibble: 15 × 6
#> plate inhibitor inhibitor_conc copies dilution n
#> <fct> <fct> <dbl> <int> <int> <int>
#> 1 soy none 0 160 625 18
#> 2 soy none 0 800 125 18
#> 3 soy none 0 4000 25 18
#> 4 soy none 0 20000 5 18
#> 5 soy none 0 100000 1 18
#> 6 soy+isopropanol isopropanol 0.004 160 625 18
#> 7 soy+isopropanol isopropanol 0.02 800 125 18
#> 8 soy+isopropanol isopropanol 0.1 4000 25 18
#> 9 soy+isopropanol isopropanol 0.5 20000 5 18
#> 10 soy+isopropanol isopropanol 2.5 100000 1 18
#> 11 soy+tannic acid tannic acid 0.0016 800 125 18
#> 12 soy+tannic acid tannic acid 0.0032 160 625 18
#> 13 soy+tannic acid tannic acid 0.008 4000 25 18
#> 14 soy+tannic acid tannic acid 0.04 20000 5 18
#> 15 soy+tannic acid tannic acid 0.2 100000 1 18
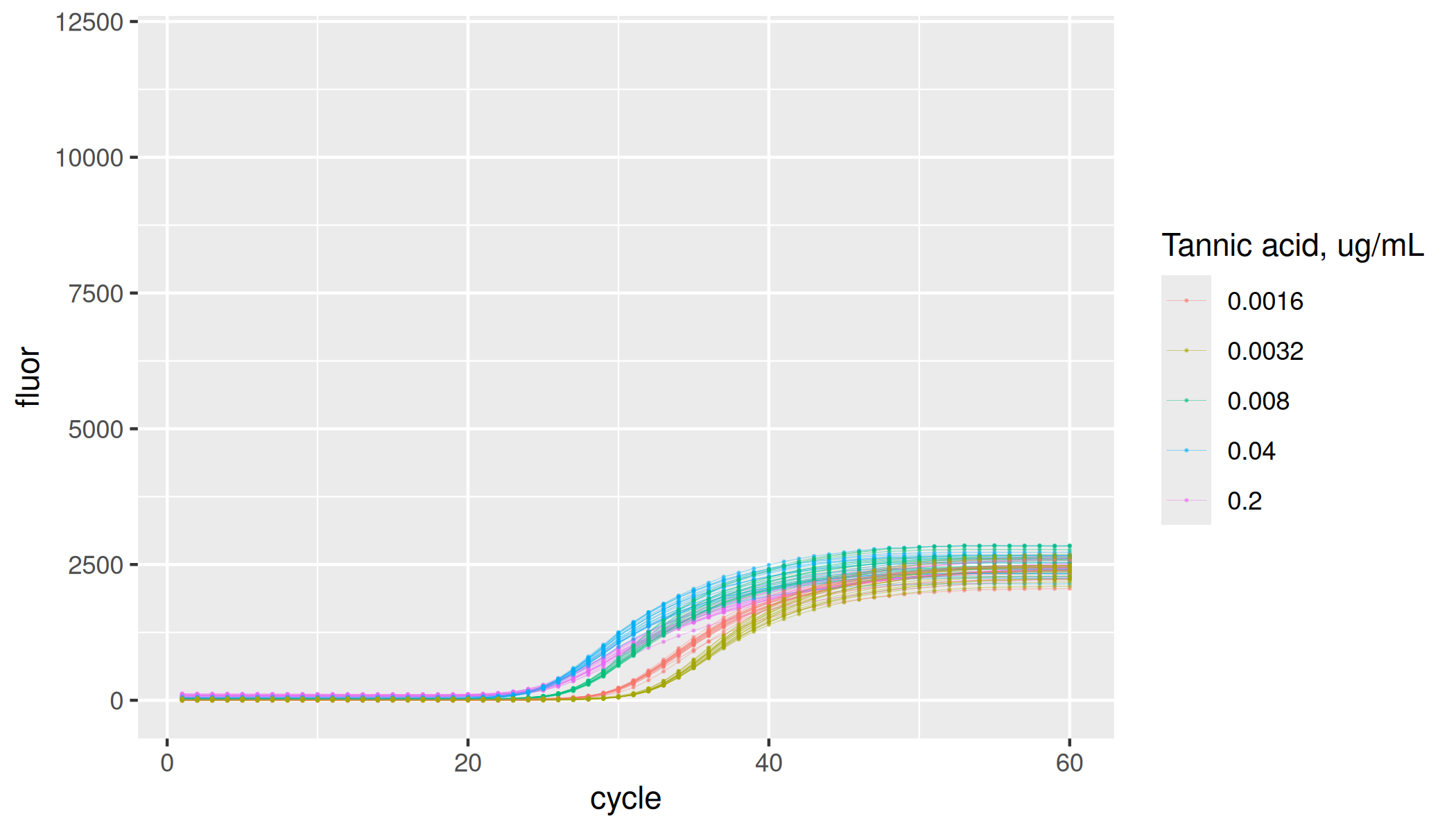
Standard dilution series
Most concentrated set of replicates in the dilution series have 100,000 copies of the soybean Lectin endogene (Le1) derived amplicon. Following samples in the series are five-fold dilutions.
lievens |>
dplyr::filter(inhibitor == "none") |>
ggplot(aes(x = cycle, y = fluor, group = interaction(plate, inhibitor, dilution, replicate), col = as.factor(dilution))) +
geom_line(linewidth = 0.1, alpha = 0.5) +
geom_point(size = 0.05, alpha = 0.5) +
labs(color = "Fold dilution") +
ylim(-100, 12000)
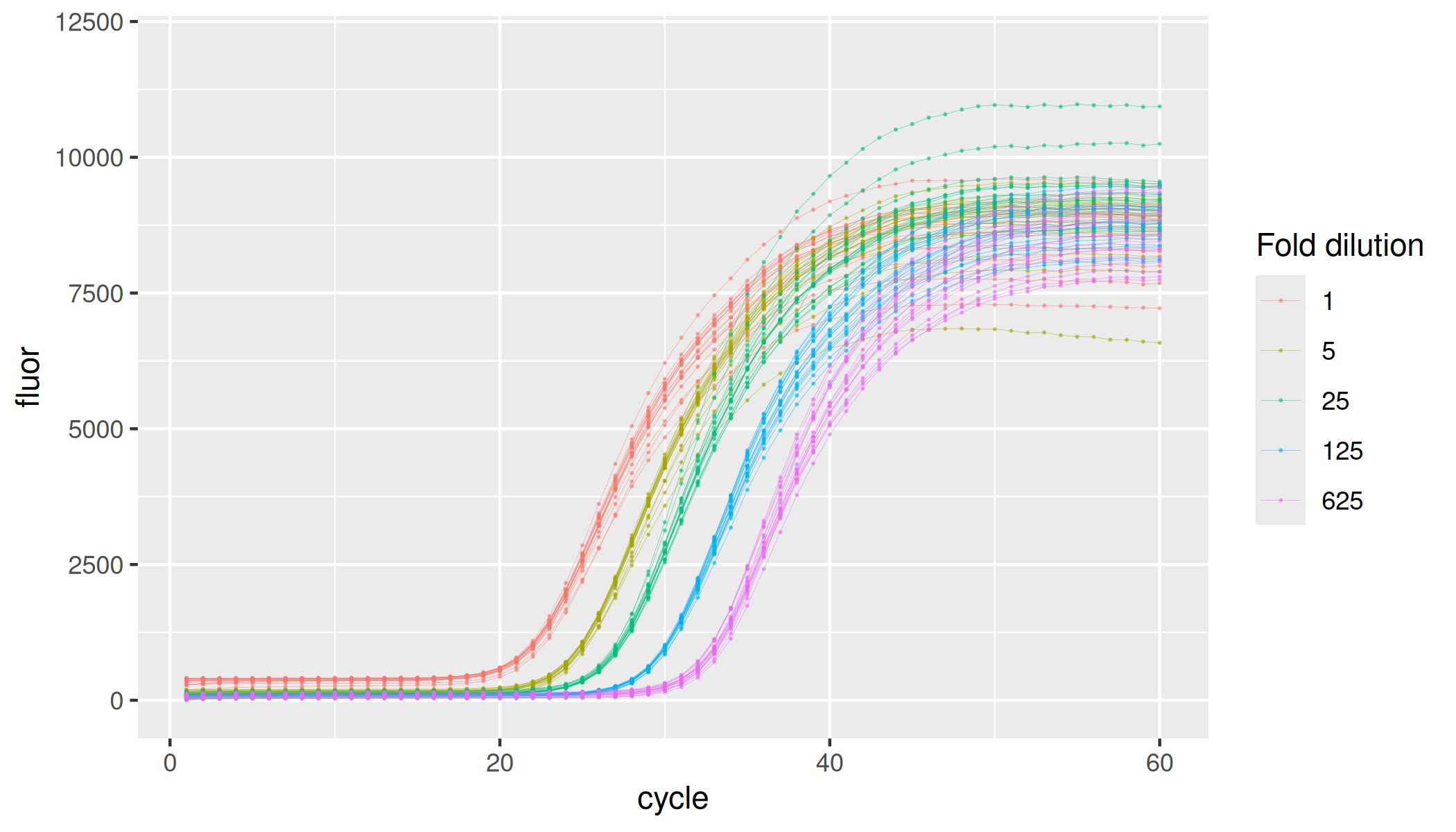
Inhibition by isopropanol
lievens |>
dplyr::filter(inhibitor == "isopropanol") |>
ggplot(aes(x = cycle, y = fluor, group = interaction(plate, inhibitor, inhibitor_conc, replicate), col = as.factor(inhibitor_conc))) +
geom_line(linewidth = 0.1, alpha = 0.5) +
geom_point(size = 0.05, alpha = 0.5) +
labs(color = "Isopropanol, % (v/v)") +
ylim(-100, 12000)
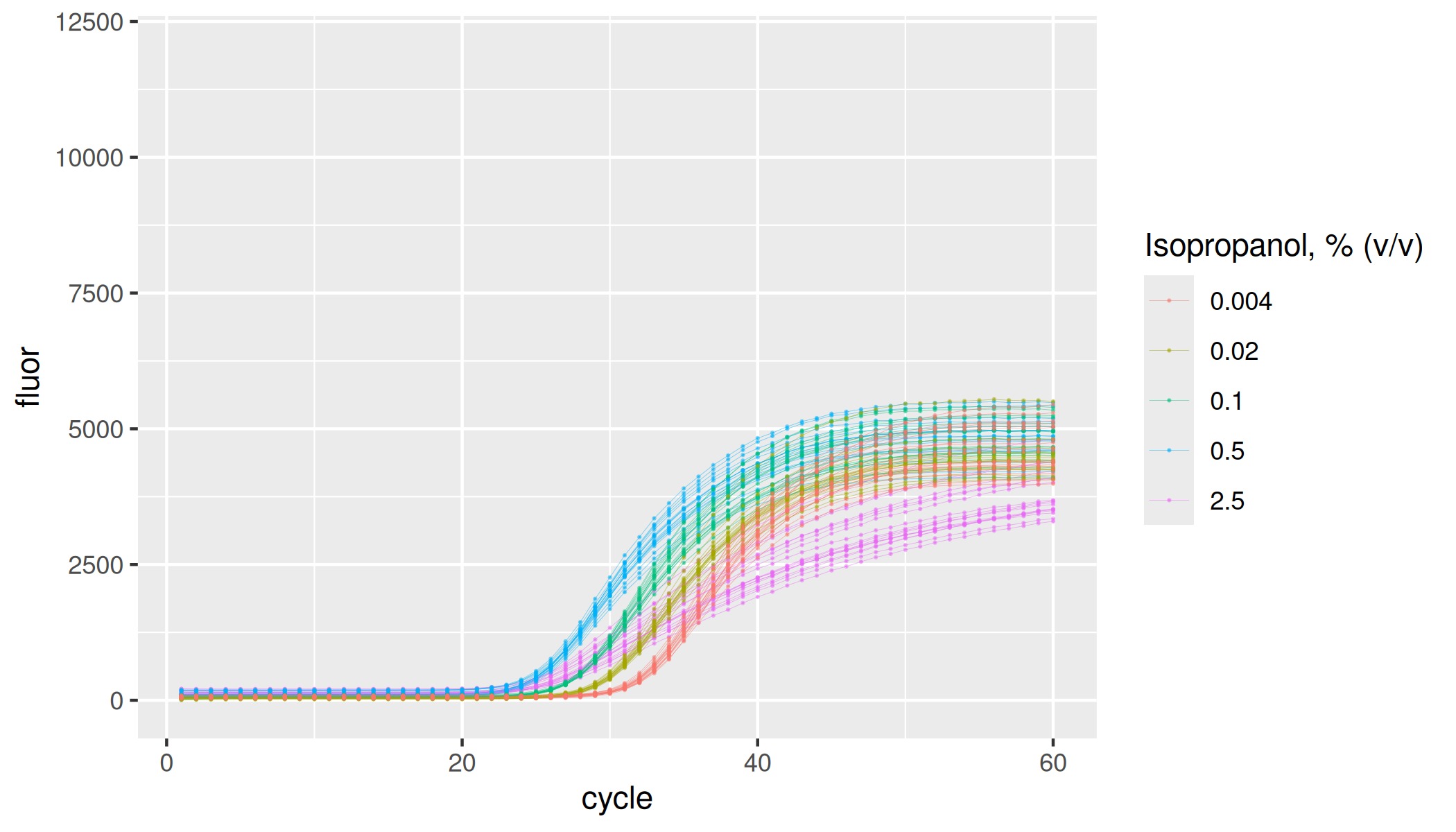
Inhibition by tannic acid
lievens |>
dplyr::filter(inhibitor == "tannic acid") |>
ggplot(aes(x = cycle, y = fluor, group = interaction(plate, inhibitor, inhibitor_conc, replicate), col = as.factor(inhibitor_conc))) +
geom_line(linewidth = 0.1, alpha = 0.5) +
geom_point(size = 0.05, alpha = 0.5) +
labs(color = "Tannic acid, ug/mL") +
ylim(-100, 12000)