Description
Veras Miscellaneous.
Description
Contains a collection of useful functions for basic data computation and manipulation, wrapper functions for generating 'ggplot2' graphics, including statistical model diagnostic plots, methods for computing statistical models quality measures (such as AIC, BIC, r squared, root mean squared error) and general utilities.
README.md
lvmisc 
lvmisc is a package with miscellaneous R functions, including basic data computation/manipulation, easy plotting and tools for working with statistical models objects. You can learn more about the methods for working with models in vignette("working_with_models").
Installation
You can install the released version of lvmisc from CRAN with:
install.packages("lvmisc")
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("verasls/lvmisc")
Getting started
Some of what you can do with lvmisc.
library(lvmisc)
library(dplyr)
# Compute body mass index (BMI) and categorize it
starwars %>%
select(name, birth_year, mass, height) %>%
mutate(
BMI = bmi(mass, height / 100),
BMI_category = bmi_cat(BMI)
)
#> # A tibble: 87 × 6
#> name birth_year mass height BMI BMI_category
#> <chr> <dbl> <dbl> <int> <dbl> <fct>
#> 1 Luke Skywalker 19 77 172 26.0 Overweight
#> 2 C-3PO 112 75 167 26.9 Overweight
#> 3 R2-D2 33 32 96 34.7 Obesity class I
#> 4 Darth Vader 41.9 136 202 33.3 Obesity class I
#> 5 Leia Organa 19 49 150 21.8 Normal weight
#> 6 Owen Lars 52 120 178 37.9 Obesity class II
#> 7 Beru Whitesun lars 47 75 165 27.5 Overweight
#> 8 R5-D4 NA 32 97 34.0 Obesity class I
#> 9 Biggs Darklighter 24 84 183 25.1 Overweight
#> 10 Obi-Wan Kenobi 57 77 182 23.2 Normal weight
#> # … with 77 more rows
# Divide numerical variables in quantiles
divide_by_quantile(mtcars$wt, 4)
#> [1] 2 2 1 2 3 3 3 2 2 3 3 4 4 4 4 4 4 1 1 1 1 3 3 4 4 1 1 1 2 2 3 2
#> Levels: 1 2 3 4
# Center and scale variables by group
center_variable(iris$Petal.Width, by = iris$Species, scale = TRUE)
#> [1] -0.046 -0.046 -0.046 -0.046 -0.046 0.154 0.054 -0.046 -0.046 -0.146
#> [11] -0.046 -0.046 -0.146 -0.146 -0.046 0.154 0.154 0.054 0.054 0.054
#> [21] -0.046 0.154 -0.046 0.254 -0.046 -0.046 0.154 -0.046 -0.046 -0.046
#> [31] -0.046 0.154 -0.146 -0.046 -0.046 -0.046 -0.046 -0.146 -0.046 -0.046
#> [41] 0.054 0.054 -0.046 0.354 0.154 0.054 -0.046 -0.046 -0.046 -0.046
#> [51] 0.074 0.174 0.174 -0.026 0.174 -0.026 0.274 -0.326 -0.026 0.074
#> [61] -0.326 0.174 -0.326 0.074 -0.026 0.074 0.174 -0.326 0.174 -0.226
#> [71] 0.474 -0.026 0.174 -0.126 -0.026 0.074 0.074 0.374 0.174 -0.326
#> [81] -0.226 -0.326 -0.126 0.274 0.174 0.274 0.174 -0.026 -0.026 -0.026
#> [91] -0.126 0.074 -0.126 -0.326 -0.026 -0.126 -0.026 -0.026 -0.226 -0.026
#> [101] 0.474 -0.126 0.074 -0.226 0.174 0.074 -0.326 -0.226 -0.226 0.474
#> [111] -0.026 -0.126 0.074 -0.026 0.374 0.274 -0.226 0.174 0.274 -0.526
#> [121] 0.274 -0.026 -0.026 -0.226 0.074 -0.226 -0.226 -0.226 0.074 -0.426
#> [131] -0.126 -0.026 0.174 -0.526 -0.626 0.274 0.374 -0.226 -0.226 0.074
#> [141] 0.374 0.274 -0.126 0.274 0.474 0.274 -0.126 -0.026 0.274 -0.226
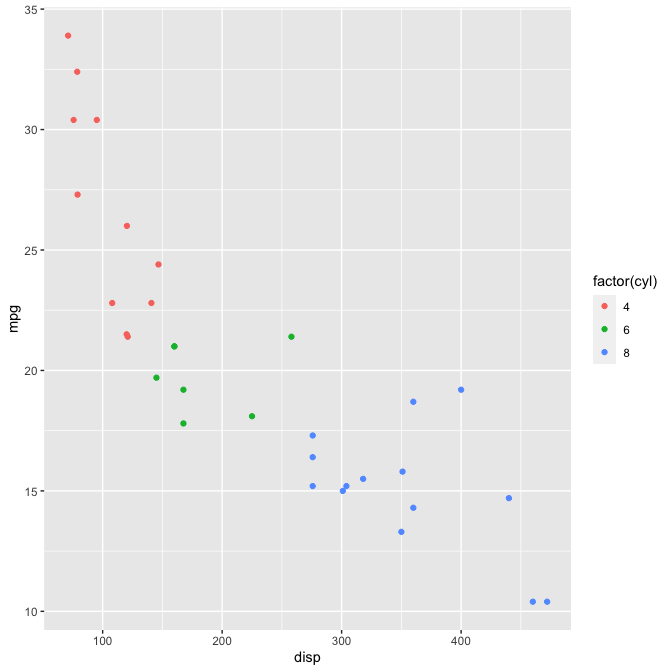
# Quick and easy plotting with {ggplot}
plot_scatter(mtcars, disp, mpg, color = factor(cyl))
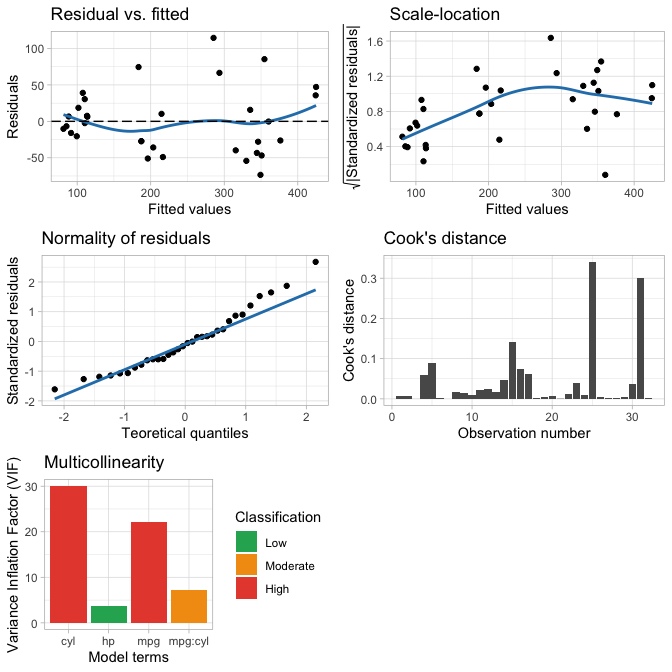
# Work with statistical model objects
m <- lm(disp ~ mpg + hp + cyl + mpg:cyl, mtcars)
accuracy(m)
#> AIC BIC R2 R2_adj MAE MAPE RMSE
#> 1 344.64 353.43 0.87 0.85 34.9 15.73% 43.75
plot_model(m)