Visualize 'Confounder' Control in Meta-Analyses.
metaconfoundr
The goal of metaconfoundr is to make it easy to visualize confounding control in meta-analyses. Researchers use a causal diagram to assess factors that are required to control for to estimate an effect unbiased by confounding. Then, the researchers create a data frame of confounding variables and how well each study in the meta-analysis controlled for each: adequately, inadequately, or with some control. See the vignette on collecting confounder data for more information.
In addition to the package, metaconfoundr ships with a Shiny app to help create visualizations. You can start the app locally with launch_metaconfoundr_app() or use the hosted version.
Installation
Install the most recent released version of metaconfoundr from CRAN:
install.packages("metaconfoundr")
You can install the development version of metaconfoundr from GitHub with:
# if needed
# install.packages("remotes")
remotes::install_github("malcolmbarrett/metaconfoundr")
Visualizing confounding control
metaconfoundr includes an example dataset, ipi, that contains information on a meta-analysis of interpregnancy interval and the risk of preterm birth. Use metaconfoundr() to prepare the dataset to work with metaconfoundr functions. ipi includes several domains of confounders, but for ease of visualization, we will just show one, Sociodemographics.
library(metaconfoundr)
mc_ipi <- metaconfoundr(ipi) %>%
dplyr::filter(construct == "Sociodemographics")
mc_ipi
#> # A tibble: 55 × 5
#> construct variable is_confounder study control_quality
#> <chr> <chr> <chr> <chr> <ord>
#> 1 Sociodemographics Maternal age Y Zhu_2001a adequate
#> 2 Sociodemographics Maternal age Y Zhu_2001b adequate
#> 3 Sociodemographics Maternal age Y Zhu_1999 adequate
#> 4 Sociodemographics Maternal age Y Smith_2003 adequate
#> 5 Sociodemographics Maternal age Y Shachar_2016 adequate
#> 6 Sociodemographics Maternal age Y Salihu_2012a adequate
#> 7 Sociodemographics Maternal age Y Salihu_2012b adequate
#> 8 Sociodemographics Maternal age Y Hanley_2017 adequate
#> 9 Sociodemographics Maternal age Y deWeger_2011 adequate
#> 10 Sociodemographics Maternal age Y Coo_2017 adequate
#> # … with 45 more rows
metaconfoundr includes several tools for visualizing the results of a confounding control assesment. The most common are mc_heatmap() and mc_trafficlight().
mc_heatmap(mc_ipi)
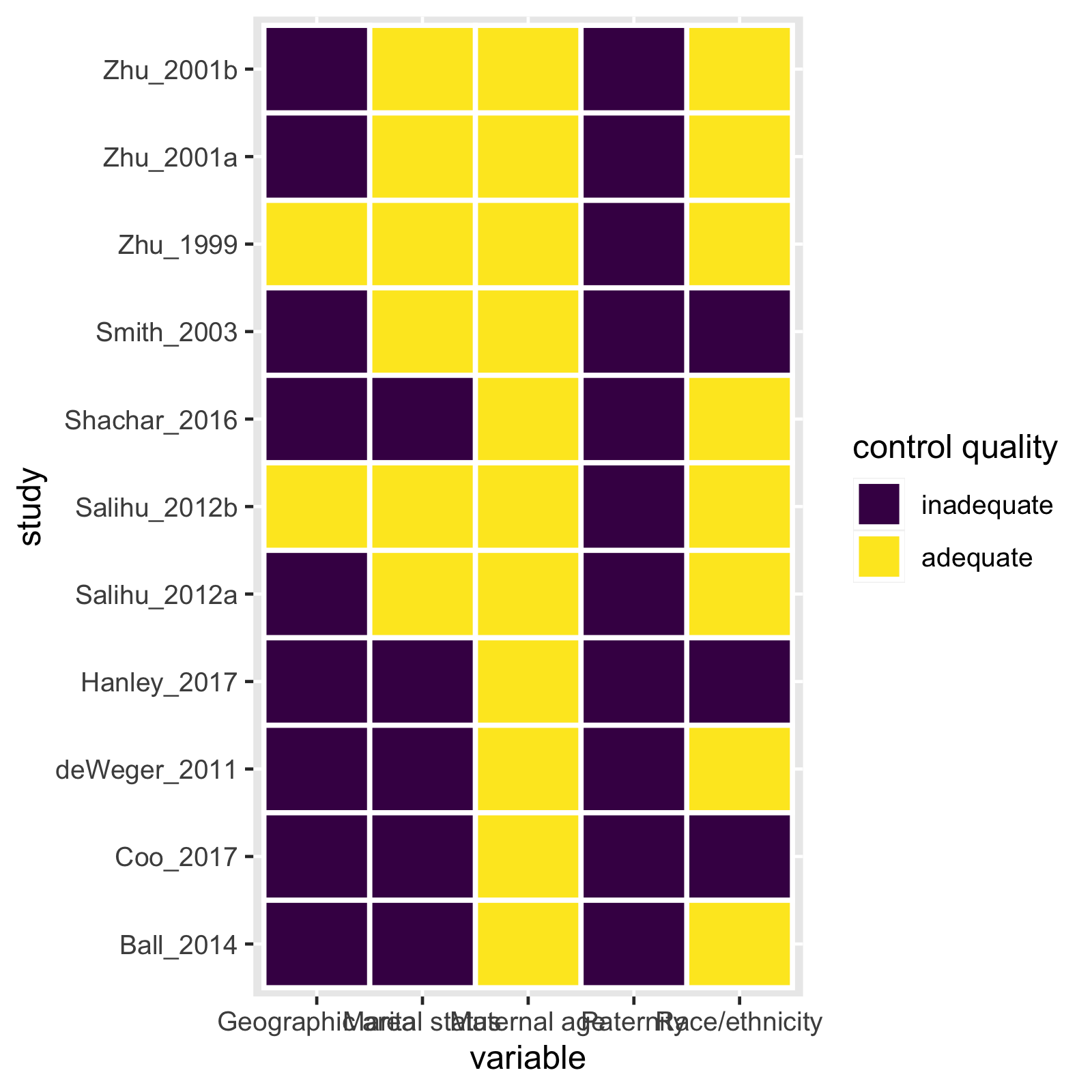
mc_trafficlight(mc_ipi)
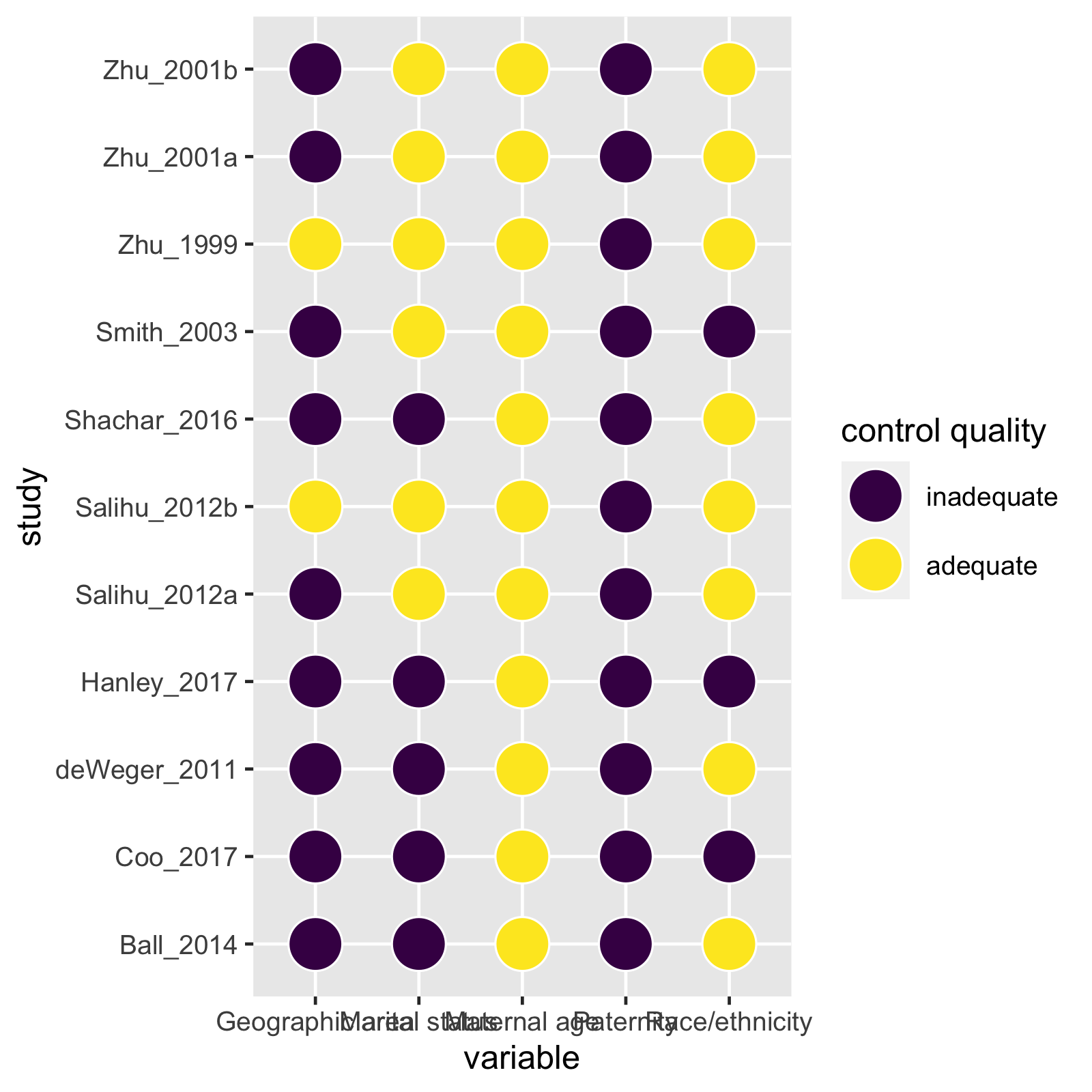
These plotting functions return ggplots, so they are easy to modify.
mc_heatmap(mc_ipi) +
theme_mc() +
ggplot2::guides(x = ggplot2::guide_axis(n.dodge = 2))
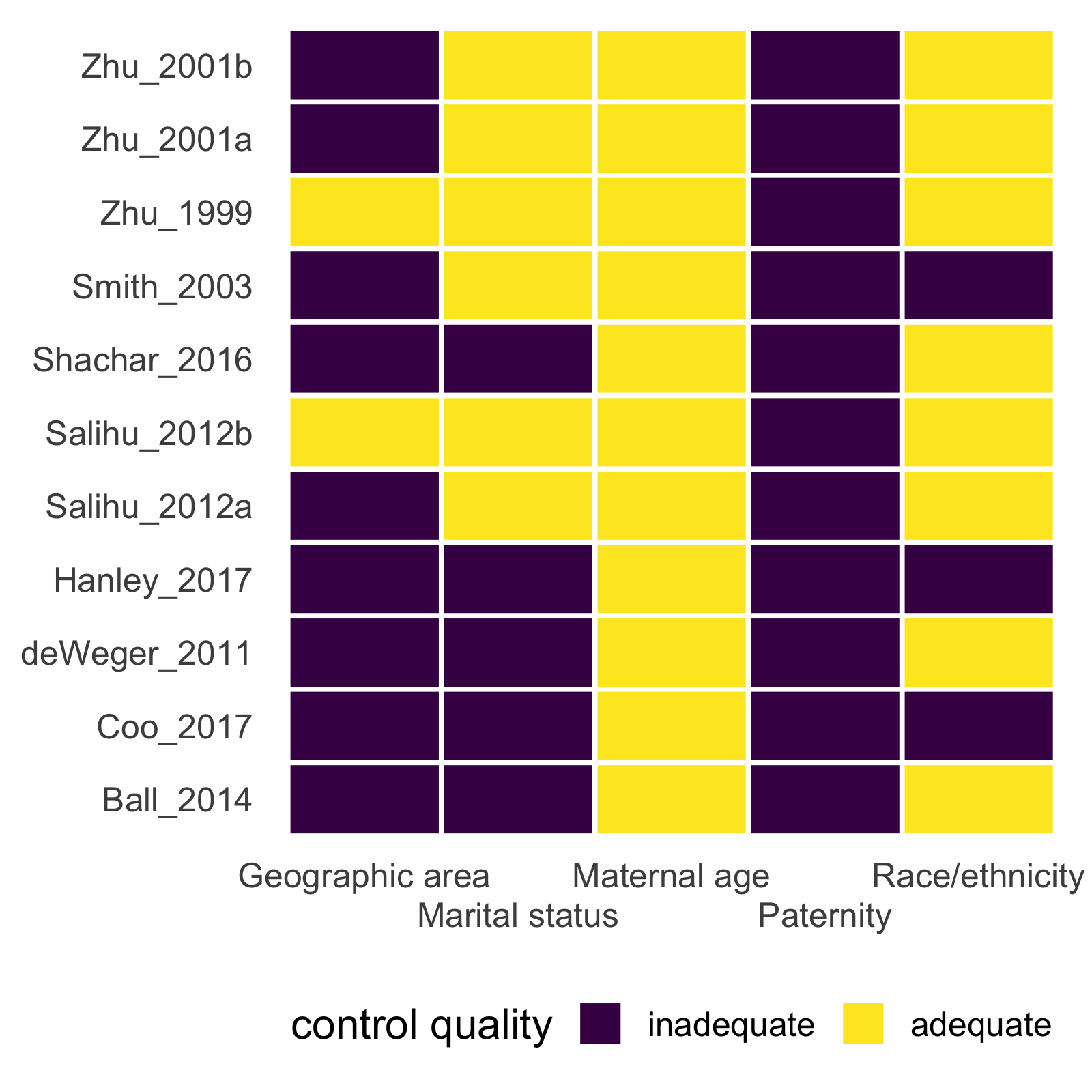
metaconfoundr also includes geoms and scales to make output similar to other types of bias assessments for meta-analyses.
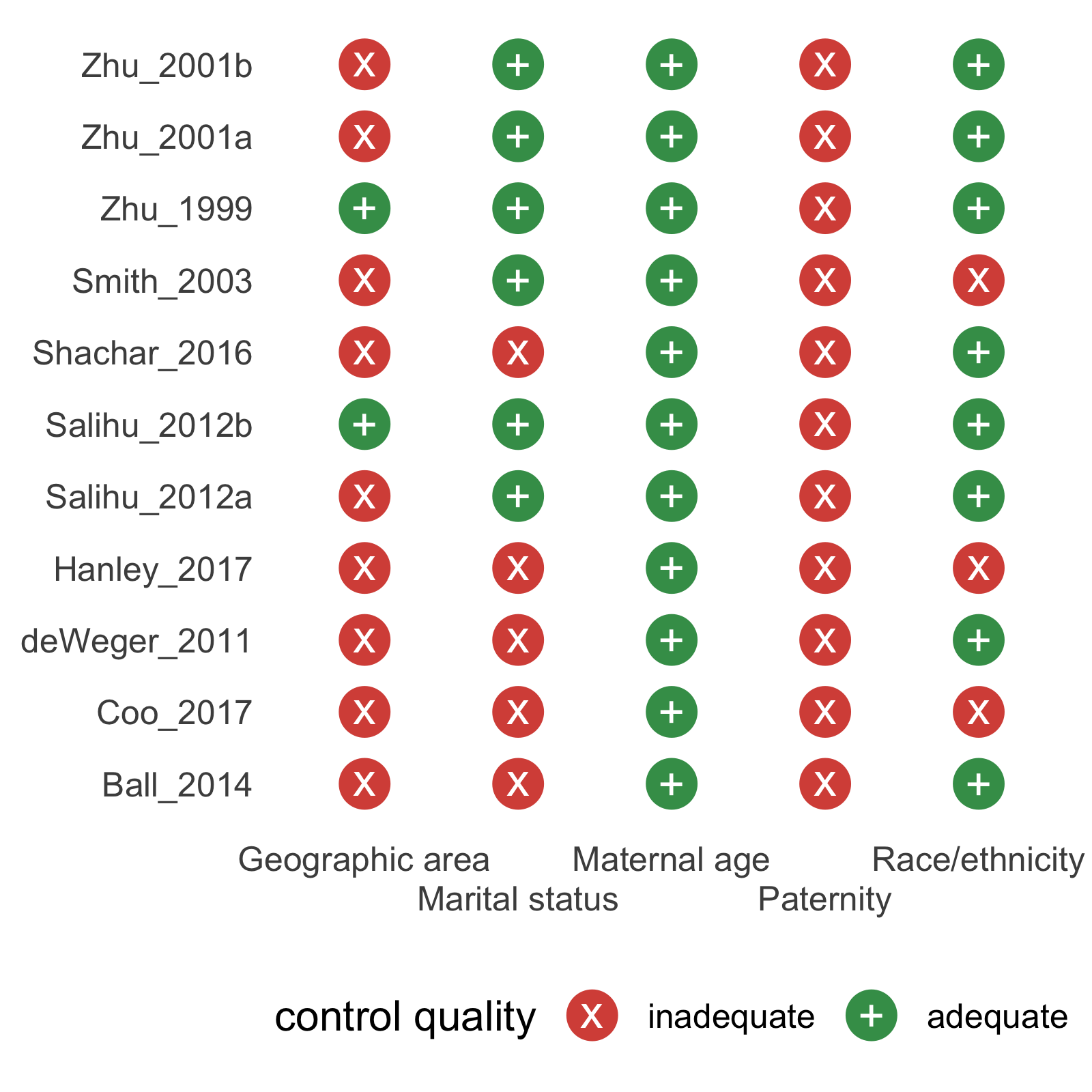
mc_trafficlight(mc_ipi) +
theme_mc() +
geom_cochrane() +
scale_fill_cochrane() +
ggplot2::guides(x = ggplot2::guide_axis(n.dodge = 2))
Similar work
For working with common assessments of bias in studies, such as the ROBINS tool, see the excellent robvis package.
Code of Conduct
Please note that the metaconfoundr project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
