Description
ADaM Metadata Structure.
Description
A metadata structure for clinical data analysis and reporting based on Analysis Data Model (ADaM) datasets. The package simplifies clinical analysis and reporting tool development by defining standardized inputs, outputs, and workflow. The package can be used to create analysis and reporting planning grid, mock table, and validated analysis and reporting results based on consistent inputs.
README.md
metalite 
Unified representation of metadata structure for clinical analysis & reporting (A&R) by leveraging the Analysis Data Model (ADaM) datasets.
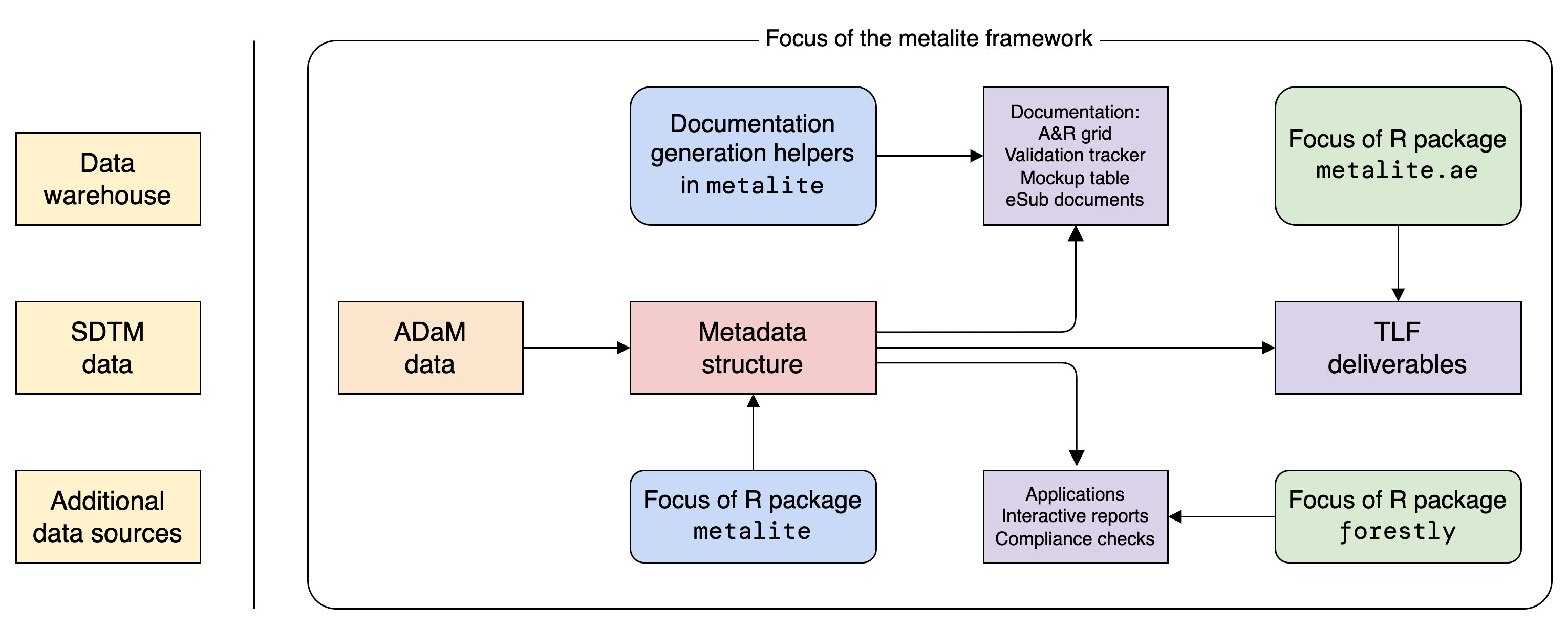
Installation
The easiest way to get metalite is to install from CRAN:
install.packages("metalite")
Alternatively, to use a new feature or get a bug fix, you can install the development version of metalite from GitHub:
# install.packages("remotes")
remotes::install_github("Merck/metalite")
Overview
The metalite framework is designed to:
- Standardize function input for analysis and reporting.
- Separate analysis logic from data source.
- Enable the use of pipes (
|>). - Reduce manual steps to develop and maintain documentation in clinical trial development.
- Ensure consistency between analysis specification, mock, and results.
Use cases
The metalite package offers a foundation to simplify tool development and create standard engineering workflows. For example, metalite can be used to:
- Standardize input and output for A&R functions.
- Create analysis and reporting planning grid.
- Create mock table.
- Create and validate A&R results.
- Trace analysis records.
Note: metalite is a low-level R package that needs to work with other R packages to complete the work. The idea is illustrated in the diagram above.
Design principles
We built metalite with the following principles:
- Automation: prefer a function call more than a checklist.
- Single-entry: enter in one place, sync to all deliveries.
- For example, enter data source one time for all AE analysis.
- End-to-end: cover all steps in software development lifecycle (SDLC) from define to delivery.