Functions and Datasets for the Book by Keon-Woong Moon.
Function "mytable"
Function "mytable"" produce table for descriptive analysis easily. It is most useful to make table to describe baseline characteristics common in medical research papers.
Basic Usage
require(moonBook)
data(acs)
mytable(Dx~.,data=acs)
Descriptive Statistics by 'Dx'
_________________________________________________________________
NSTEMI STEMI Unstable Angina p
(N=153) (N=304) (N=400)
-----------------------------------------------------------------
age 64.3 ± 12.3 62.1 ± 12.1 63.8 ± 11.0 0.073
sex 0.012
- Female 50 (32.7%) 84 (27.6%) 153 (38.2%)
- Male 103 (67.3%) 220 (72.4%) 247 (61.8%)
cardiogenicShock 0.000
- No 149 (97.4%) 256 (84.2%) 400 (100.0%)
- Yes 4 ( 2.6%) 48 (15.8%) 0 ( 0.0%)
entry 0.001
- Femoral 58 (37.9%) 133 (43.8%) 121 (30.2%)
- Radial 95 (62.1%) 171 (56.2%) 279 (69.8%)
EF 55.0 ± 9.3 52.4 ± 9.5 59.2 ± 8.7 0.000
height 163.3 ± 8.2 165.1 ± 8.2 161.7 ± 9.7 0.000
weight 64.3 ± 10.2 65.7 ± 11.6 64.5 ± 11.6 0.361
BMI 24.1 ± 3.2 24.0 ± 3.3 24.6 ± 3.4 0.064
obesity 0.186
- No 106 (69.3%) 209 (68.8%) 252 (63.0%)
- Yes 47 (30.7%) 95 (31.2%) 148 (37.0%)
TC 193.7 ± 53.6 183.2 ± 43.4 183.5 ± 48.3 0.057
LDLC 126.1 ± 44.7 116.7 ± 39.5 112.9 ± 40.4 0.004
HDLC 38.9 ± 11.9 38.5 ± 11.0 37.8 ± 10.9 0.501
TG 130.1 ± 88.5 106.5 ± 72.0 137.4 ± 101.6 0.000
DM 0.209
- No 96 (62.7%) 208 (68.4%) 249 (62.2%)
- Yes 57 (37.3%) 96 (31.6%) 151 (37.8%)
HBP 0.002
- No 62 (40.5%) 150 (49.3%) 144 (36.0%)
- Yes 91 (59.5%) 154 (50.7%) 256 (64.0%)
smoking 0.000
- Ex-smoker 42 (27.5%) 66 (21.7%) 96 (24.0%)
- Never 50 (32.7%) 97 (31.9%) 185 (46.2%)
- Smoker 61 (39.9%) 141 (46.4%) 119 (29.8%)
-----------------------------------------------------------------
The first argument of function mytable is an object of class formula. Left side of ~ must contain the name of one grouping variable or two grouping variables in an additive way(e.g. sex+group~), and the right side of ~ must have variables in an additive way. . is allowed on the right side of formula which means all variables in the data.frame specified by the 2nd argument data. The sample data 'acs' containing demographic data and laboratory data of 857 patients with acute coronary syndrome(ACS). For more information about the data acs, type ?acs in your R console.
str(acs)
'data.frame': 857 obs. of 17 variables:
$ age : int 62 78 76 89 56 73 58 62 59 71 ...
$ sex : chr "Male" "Female" "Female" "Female" ...
$ cardiogenicShock: chr "No" "No" "Yes" "No" ...
$ entry : chr "Femoral" "Femoral" "Femoral" "Femoral" ...
$ Dx : chr "STEMI" "STEMI" "STEMI" "STEMI" ...
$ EF : num 18 18.4 20 21.8 21.8 22 24.7 26.6 28.5 31.1 ...
$ height : num 168 148 NA 165 162 153 167 160 152 168 ...
$ weight : num 72 48 NA 50 64 59 78 50 67 60 ...
$ BMI : num 25.5 21.9 NA 18.4 24.4 ...
$ obesity : chr "Yes" "No" "No" "No" ...
$ TC : num 215 NA NA 121 195 184 161 136 239 169 ...
$ LDLC : int 154 NA NA 73 151 112 91 88 161 88 ...
$ HDLC : int 35 NA NA 20 36 38 34 33 34 54 ...
$ TG : int 155 166 NA 89 63 137 196 30 118 141 ...
$ DM : chr "Yes" "No" "No" "No" ...
$ HBP : chr "No" "Yes" "Yes" "No" ...
$ smoking : chr "Smoker" "Never" "Never" "Never" ...
Choosing grouping variable(s) and row-variable(s)
You can choose the grouping variable(s) and row-variable(s) with the formula.
mytable(sex~age+Dx,data=acs)
Descriptive Statistics by 'sex'
__________________________________________________
Female Male p
(N=287) (N=570)
--------------------------------------------------
age 68.7 ± 10.7 60.6 ± 11.2 0.000
Dx 0.012
- NSTEMI 50 (17.4%) 103 (18.1%)
- STEMI 84 (29.3%) 220 (38.6%)
- Unstable Angina 153 (53.3%) 247 (43.3%)
--------------------------------------------------
You can choose row-variable(s) with . and + and - and variable name in an additive way.
mytable(am~.-hp-disp-cyl-carb-gear,data=mtcars)
Descriptive Statistics by 'am'
____________________________________
0 1 p
(N=19) (N=13)
------------------------------------
mpg 17.1 ± 3.8 24.4 ± 6.2 0.000
drat 3.3 ± 0.4 4.0 ± 0.4 0.000
wt 3.8 ± 0.8 2.4 ± 0.6 0.000
qsec 18.2 ± 1.8 17.4 ± 1.8 0.206
vs 0.556
- 0 12 (63.2%) 6 (46.2%)
- 1 7 (36.8%) 7 (53.8%)
------------------------------------
Method for continuous variables
By default continuous variables are analyzed as normal-distributed and are described with mean and standard deviation. To change default options, you can use the method argument. Possible values of method argument are:
- 1: forces analysis as normal-distributed, default value
- 2: forces analysis as continuous non-normal
- 3: performs a Shapiro-Wilks test to decide between normal or non-normal
When continuous variables are analyzed as non-normal, they are described with median and interquartile range.
mytable(sex~height+weight+BMI,data=acs,method=3)
Descriptive Statistics by 'sex'
_____________________________________________________
Female Male p
(N=287) (N=570)
-----------------------------------------------------
height 155.0 [150.0;158.0] 168.0 [164.0;172.0] 0.000
weight 58.0 [50.0;63.0] 68.0 [62.0;75.0] 0.000
BMI 24.0 [22.1;26.2] 24.2 [22.2;26.2] 0.471
-----------------------------------------------------
Because the method argument is selected as 3, a Shapiro-Wilk test normality test is used to decide if the variable is normal or non-normal distributed. Note that height and BMI was described as mean $\pm$ sd, whereas the weight was described as median and interquartile range.
choice of variable : categorical or continuous variable - my way
In many cases, categorical variables are usually coded as numeric. For example, many people usually code 0 and 1 instead of "No" and "Yes". Similarly, factor variables with three or four levels are coded 0/1/2 or 0/1/2/3. In many cases, if we analyze these variables as continuous variables, we are not able to get the right result. In mytable, variables with less than five unique values are treated as a categorical variables.
mytable(am~.,data=mtcars)
Descriptive Statistics by 'am'
_______________________________________
0 1 p
(N=19) (N=13)
---------------------------------------
mpg 17.1 ± 3.8 24.4 ± 6.2 0.000
cyl 0.013
- 4 3 (15.8%) 8 (61.5%)
- 6 4 (21.1%) 3 (23.1%)
- 8 12 (63.2%) 2 (15.4%)
disp 290.4 ± 110.2 143.5 ± 87.2 0.000
hp 160.3 ± 53.9 126.8 ± 84.1 0.180
drat 3.3 ± 0.4 4.0 ± 0.4 0.000
wt 3.8 ± 0.8 2.4 ± 0.6 0.000
qsec 18.2 ± 1.8 17.4 ± 1.8 0.206
vs 0.556
- 0 12 (63.2%) 6 (46.2%)
- 1 7 (36.8%) 7 (53.8%)
gear 0.000
- 3 15 (78.9%) 0 ( 0.0%)
- 4 4 (21.1%) 8 (61.5%)
- 5 0 ( 0.0%) 5 (38.5%)
carb 2.7 ± 1.1 2.9 ± 2.2 0.781
---------------------------------------
In mtcars data, all variables are expressed as numeric. But as you can see, cyl, vs and gear is treated as categorical variables. The carb variables has six unique values and treated as continuous variables. If you wanted the carb variable to be treated as categorical variable, you can changed the max.ylev argument.
mytable(am~carb,data=mtcars,max.ylev=6)
Descriptive Statistics by 'am'
__________________________________
0 1 p
(N=19) (N=13)
----------------------------------
carb 0.284
- 1 3 (15.8%) 4 (30.8%)
- 2 6 (31.6%) 4 (30.8%)
- 3 3 (15.8%) 0 ( 0.0%)
- 4 7 (36.8%) 3 (23.1%)
- 6 0 ( 0.0%) 1 ( 7.7%)
- 8 0 ( 0.0%) 1 ( 7.7%)
----------------------------------
Combining tables
If you wanted to make two separate tables and combine into one table, mytable is the function of choice. For example, if you wanted to build separate table for female and male patients stratified by presence or absence of DM and combine it,
mytable(sex+DM~.,data=acs)
Descriptive Statistics stratified by 'sex' and 'DM'
_____________________________________________________________________________________
Male Female
-------------------------------- -------------------------------
No Yes p No Yes p
(N=380) (N=190) (N=173) (N=114)
-------------------------------------------------------------------------------------
age 60.9 ± 11.5 60.1 ± 10.6 0.472 69.3 ± 11.4 67.8 ± 9.7 0.257
cardiogenicShock 0.685 0.296
- No 355 (93.4%) 175 (92.1%) 168 (97.1%) 107 (93.9%)
- Yes 25 ( 6.6%) 15 ( 7.9%) 5 ( 2.9%) 7 ( 6.1%)
entry 0.552 0.665
- Femoral 125 (32.9%) 68 (35.8%) 74 (42.8%) 45 (39.5%)
- Radial 255 (67.1%) 122 (64.2%) 99 (57.2%) 69 (60.5%)
Dx 0.219 0.240
- NSTEMI 71 (18.7%) 32 (16.8%) 25 (14.5%) 25 (21.9%)
- STEMI 154 (40.5%) 66 (34.7%) 54 (31.2%) 30 (26.3%)
- Unstable Angina 155 (40.8%) 92 (48.4%) 94 (54.3%) 59 (51.8%)
EF 56.5 ± 8.3 53.9 ± 11.0 0.007 56.0 ± 10.1 56.6 ± 10.0 0.655
height 168.1 ± 5.8 167.5 ± 6.7 0.386 153.9 ± 6.5 153.6 ± 5.8 0.707
weight 68.1 ± 10.4 69.8 ± 10.2 0.070 56.5 ± 8.7 58.4 ± 10.0 0.106
BMI 24.0 ± 3.1 24.9 ± 3.5 0.005 23.8 ± 3.2 24.8 ± 4.0 0.046
obesity 0.027 0.359
- No 261 (68.7%) 112 (58.9%) 121 (69.9%) 73 (64.0%)
- Yes 119 (31.3%) 78 (41.1%) 52 (30.1%) 41 (36.0%)
TC 184.1 ± 46.7 181.8 ± 44.5 0.572 186.0 ± 43.1 193.3 ± 60.8 0.274
LDLC 117.9 ± 41.8 112.1 ± 39.4 0.115 116.3 ± 35.2 119.8 ± 48.6 0.519
HDLC 38.4 ± 11.4 36.8 ± 9.6 0.083 39.2 ± 10.9 38.8 ± 12.2 0.821
TG 115.2 ± 72.2 153.4 ± 130.7 0.000 114.2 ± 82.4 128.4 ± 65.5 0.112
HBP 0.000 0.356
- No 205 (53.9%) 68 (35.8%) 54 (31.2%) 29 (25.4%)
- Yes 175 (46.1%) 122 (64.2%) 119 (68.8%) 85 (74.6%)
smoking 0.386 0.093
- Ex-smoker 101 (26.6%) 54 (28.4%) 34 (19.7%) 15 (13.2%)
- Never 77 (20.3%) 46 (24.2%) 118 (68.2%) 91 (79.8%)
- Smoker 202 (53.2%) 90 (47.4%) 21 (12.1%) 8 ( 7.0%)
-------------------------------------------------------------------------------------
For more beautiful output : myhtml
If you want more beautiful table in your R markdown file, you can use myhtml function.
out=mytable(Dx~.,data=acs)
myhtml(out)
| Dx | NSTEMI (N=153) | STEMI (N=304) | Unstable Angina (N=400) | p |
|---|---|---|---|---|
| age | 64.3 ± 12.3 | 62.1 ± 12.1 | 63.8 ± 11.0 | 0.073 |
| sex | 0.012 | |||
| Female | 50 (32.7%) | 84 (27.6%) | 153 (38.2%) | |
| Male | 103 (67.3%) | 220 (72.4%) | 247 (61.8%) | |
| cardiogenicShock | 0.000 | |||
| No | 149 (97.4%) | 256 (84.2%) | 400 (100.0%) | |
| Yes | 4 ( 2.6%) | 48 (15.8%) | 0 ( 0.0%) | |
| entry | 0.001 | |||
| Femoral | 58 (37.9%) | 133 (43.8%) | 121 (30.2%) | |
| Radial | 95 (62.1%) | 171 (56.2%) | 279 (69.8%) | |
| EF | 55.0 ± 9.3 | 52.4 ± 9.5 | 59.2 ± 8.7 | 0.000 |
| height | 163.3 ± 8.2 | 165.1 ± 8.2 | 161.7 ± 9.7 | 0.000 |
| weight | 64.3 ± 10.2 | 65.7 ± 11.6 | 64.5 ± 11.6 | 0.361 |
| BMI | 24.1 ± 3.2 | 24.0 ± 3.3 | 24.6 ± 3.4 | 0.064 |
| obesity | 0.186 | |||
| No | 106 (69.3%) | 209 (68.8%) | 252 (63.0%) | |
| Yes | 47 (30.7%) | 95 (31.2%) | 148 (37.0%) | |
| TC | 193.7 ± 53.6 | 183.2 ± 43.4 | 183.5 ± 48.3 | 0.057 |
| LDLC | 126.1 ± 44.7 | 116.7 ± 39.5 | 112.9 ± 40.4 | 0.004 |
| HDLC | 38.9 ± 11.9 | 38.5 ± 11.0 | 37.8 ± 10.9 | 0.501 |
| TG | 130.1 ± 88.5 | 106.5 ± 72.0 | 137.4 ± 101.6 | 0.000 |
| DM | 0.209 | |||
| No | 96 (62.7%) | 208 (68.4%) | 249 (62.2%) | |
| Yes | 57 (37.3%) | 96 (31.6%) | 151 (37.8%) | |
| HBP | 0.002 | |||
| No | 62 (40.5%) | 150 (49.3%) | 144 (36.0%) | |
| Yes | 91 (59.5%) | 154 (50.7%) | 256 (64.0%) | |
| smoking | 0.000 | |||
| Ex-smoker | 42 (27.5%) | 66 (21.7%) | 96 (24.0%) | |
| Never | 50 (32.7%) | 97 (31.9%) | 185 (46.2%) | |
| Smoker | 61 (39.9%) | 141 (46.4%) | 119 (29.8%) |
out1=mytable(sex+DM~.,data=acs)
myhtml(out1)
| sex | Male | Female | ||||
|---|---|---|---|---|---|---|
| DM | No (N=380) | Yes (N=190) | p | No (N=173) | Yes (N=114) | p |
| age | 60.9 ± 11.5 | 60.1 ± 10.6 | 0.472 | 69.3 ± 11.4 | 67.8 ± 9.7 | 0.257 |
| cardiogenicShock | 0.685 | 0.296 | ||||
| No | 355 (93.4%) | 175 (92.1%) | 168 (97.1%) | 107 (93.9%) | ||
| Yes | 25 ( 6.6%) | 15 ( 7.9%) | 5 ( 2.9%) | 7 ( 6.1%) | ||
| entry | 0.552 | 0.665 | ||||
| Femoral | 125 (32.9%) | 68 (35.8%) | 74 (42.8%) | 45 (39.5%) | ||
| Radial | 255 (67.1%) | 122 (64.2%) | 99 (57.2%) | 69 (60.5%) | ||
| Dx | 0.219 | 0.240 | ||||
| NSTEMI | 71 (18.7%) | 32 (16.8%) | 25 (14.5%) | 25 (21.9%) | ||
| STEMI | 154 (40.5%) | 66 (34.7%) | 54 (31.2%) | 30 (26.3%) | ||
| Unstable Angina | 155 (40.8%) | 92 (48.4%) | 94 (54.3%) | 59 (51.8%) | ||
| EF | 56.5 ± 8.3 | 53.9 ± 11.0 | 0.007 | 56.0 ± 10.1 | 56.6 ± 10.0 | 0.655 |
| height | 168.1 ± 5.8 | 167.5 ± 6.7 | 0.386 | 153.9 ± 6.5 | 153.6 ± 5.8 | 0.707 |
| weight | 68.1 ± 10.4 | 69.8 ± 10.2 | 0.070 | 56.5 ± 8.7 | 58.4 ± 10.0 | 0.106 |
| BMI | 24.0 ± 3.1 | 24.9 ± 3.5 | 0.005 | 23.8 ± 3.2 | 24.8 ± 4.0 | 0.046 |
| obesity | 0.027 | 0.359 | ||||
| No | 261 (68.7%) | 112 (58.9%) | 121 (69.9%) | 73 (64.0%) | ||
| Yes | 119 (31.3%) | 78 (41.1%) | 52 (30.1%) | 41 (36.0%) | ||
| TC | 184.1 ± 46.7 | 181.8 ± 44.5 | 0.572 | 186.0 ± 43.1 | 193.3 ± 60.8 | 0.274 |
| LDLC | 117.9 ± 41.8 | 112.1 ± 39.4 | 0.115 | 116.3 ± 35.2 | 119.8 ± 48.6 | 0.519 |
| HDLC | 38.4 ± 11.4 | 36.8 ± 9.6 | 0.083 | 39.2 ± 10.9 | 38.8 ± 12.2 | 0.821 |
| TG | 115.2 ± 72.2 | 153.4 ± 130.7 | 0.000 | 114.2 ± 82.4 | 128.4 ± 65.5 | 0.112 |
| HBP | 0.000 | 0.356 | ||||
| No | 205 (53.9%) | 68 (35.8%) | 54 (31.2%) | 29 (25.4%) | ||
| Yes | 175 (46.1%) | 122 (64.2%) | 119 (68.8%) | 85 (74.6%) | ||
| smoking | 0.386 | 0.093 | ||||
| Ex-smoker | 101 (26.6%) | 54 (28.4%) | 34 (19.7%) | 15 (13.2%) | ||
| Never | 77 (20.3%) | 46 (24.2%) | 118 (68.2%) | 91 (79.8%) | ||
| Smoker | 202 (53.2%) | 90 (47.4%) | 21 (12.1%) | 8 ( 7.0%) | ||
For more beautiful output : mylatex
If you want more beautiful table, you can use mylatex function.
mylatex(mytable(sex+DM~age+Dx,data=acs))
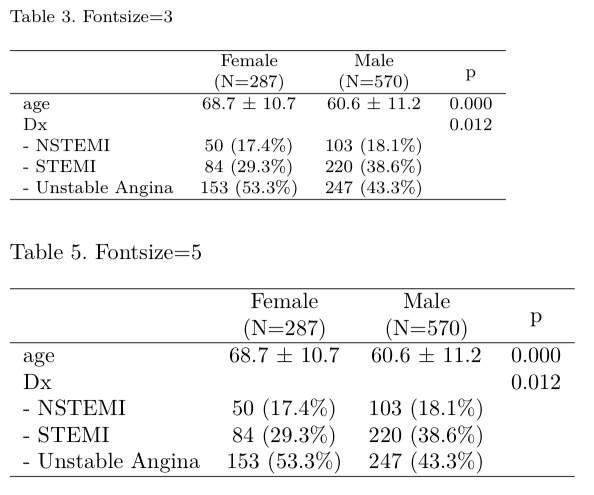
You can adjust font size of latex table by using parameter size from 1 to 10.
out=mytable(sex~age+Dx,data=acs)
for(i in c(3,5))
mylatex(out,size=i,caption=paste("Table ",i,". Fontsize=",i,sep=""))

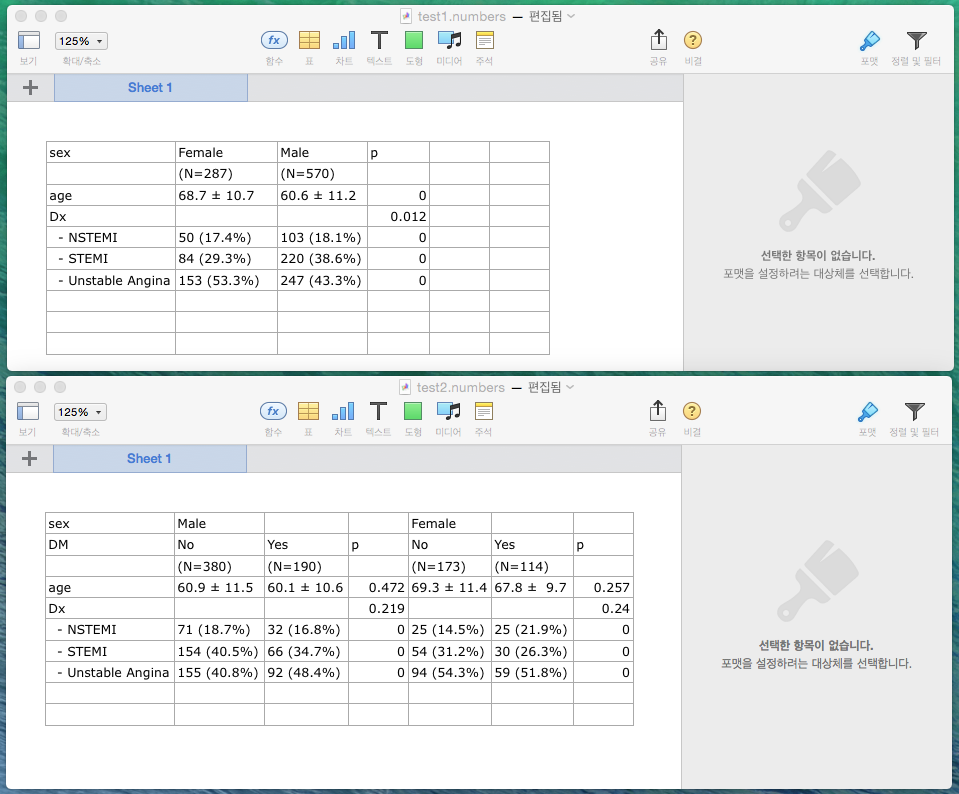
Export to csv file : mycsv
If you want to export your table into csv file format, you can use mycsv function.
mycsv(out,file="test.csv")
mycsv(out1,fil="test1.csv")
Following figure is a screen-shot in which test.csv and test1.csv files are opened with Numbers.
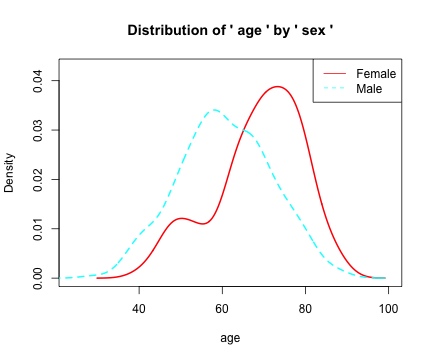
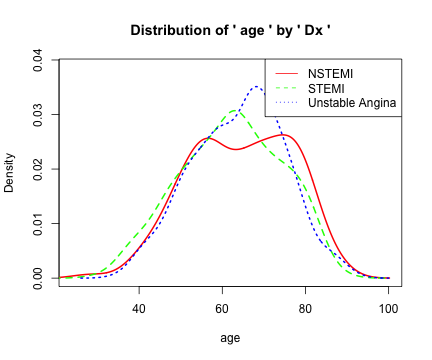
densityplot
library(moonBook)
densityplot(age~sex,data=acs)
densityplot(age~Dx,data=acs)
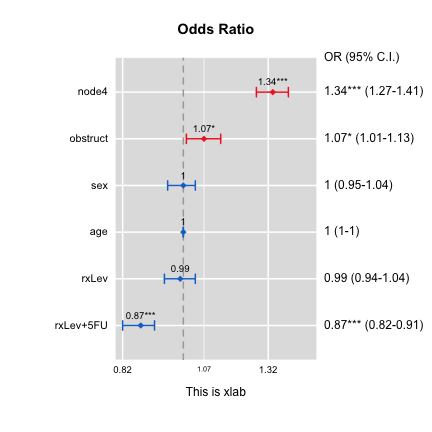
Plot for odds ratios of a glm object
require(survival)
data(colon)
out1=glm(status~sex+age+rx+obstruct+node4,data=colon)
out2=glm(status~rx+node4,data=colon)
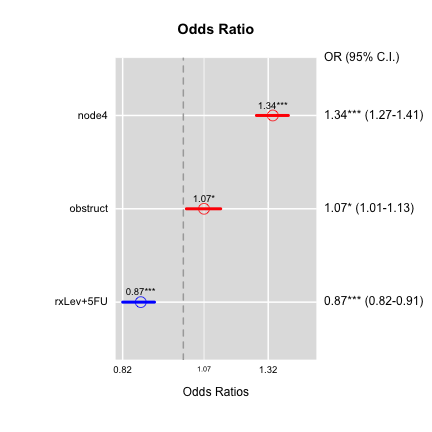
ORplot(out1,type=2,show.CI=TRUE,xlab="This is xlab",main="Odds Ratio")
ORplot(out2,type=1)
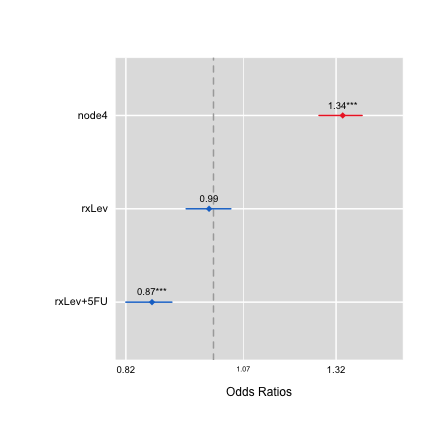
ORplot(out1,type=1,show.CI=TRUE,col=c("blue","red"))
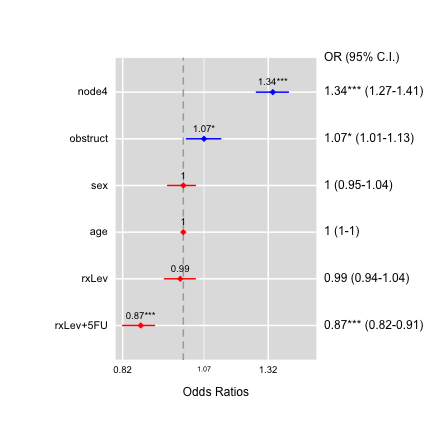
ORplot(out1,type=4,show.CI=TRUE,sig.level=0.05)
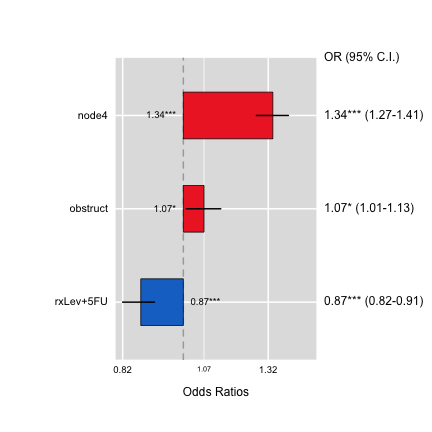
ORplot(out1,type=1,show.CI=TRUE,main="Odds Ratio",sig.level=0.05,
pch=1,cex=2,lwd=4,col=c("red","blue"))
For automation of cox's proportional hazard model
attach(colon)
The following objects are masked from colon (pos = 4):
adhere, age, differ, etype, extent, id, node4, nodes,
obstruct, perfor, rx, sex, status, study, surg, time
colon$TS=Surv(time,status==1)
out=mycph(TS~.,data=colon)
mycph : perform coxph of individual expecting variables
Call: TS ~ ., data= colon
study was excluded : NaN
status was excluded : infinite
out
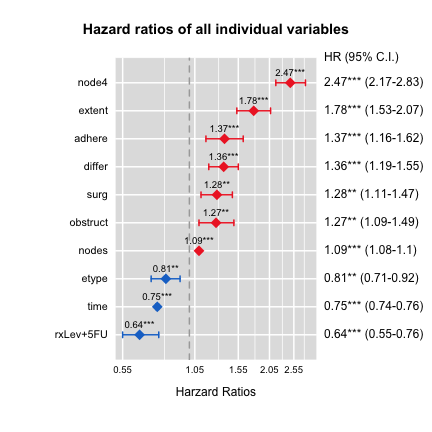
HR lcl ucl p
id 1.00 1.00 1.00 0.317
rxLev 0.98 0.84 1.14 0.786
rxLev+5FU 0.64 0.55 0.76 0.000
sex 0.97 0.85 1.10 0.610
age 1.00 0.99 1.00 0.382
obstruct 1.27 1.09 1.49 0.003
perfor 1.30 0.92 1.85 0.142
adhere 1.37 1.16 1.62 0.000
nodes 1.09 1.08 1.10 0.000
differ 1.36 1.19 1.55 0.000
extent 1.78 1.53 2.07 0.000
surg 1.28 1.11 1.47 0.001
node4 2.47 2.17 2.83 0.000
time 0.75 0.74 0.76 0.000
etype 0.81 0.71 0.92 0.001
HRplot(out,type=2,show.CI=TRUE,cex=2,sig=0.05,
main="Hazard ratios of all individual variables")