Multidimensional Top Scoring for Creativity Research.
Multidimensional Top Scoring for Creativity Research 
An R adaptation of Multidimensional Top Scoring method presented by Forthmann, Karwowski and Beaty (2023) using the code from the OSF database. The code was adapted to use the tidyverse framework for greater flexibility.
Installation
Install mtscr with:
install.packages("mtscr")
You can install the development version of mtscr from GitHub with:
# install.packages("devtools")
devtools::install_github("jakub-jedrusiak/mtscr")
Usage
Basic usage involves scoring participants’ responses to a divergent thinking task. The package includes a sample dataset mtscr_creativity with 4652 responses to the Alternative Uses Task with semantic distance scored. The dataset comes from the original paper (Forthmann, Karwowski and Beaty, 2023).
The main function is mtscr_scores() which returns a dataframe with scored responses. It takes a dataframe with responses, an ID column, an item column and a score column as arguments. The score column should contain semantic distance scores for each response. The function adds columns with scores for each person. The number of creativity scores is based on a given number of top answers provided by the top argument.
library("mtscr")
data("mtscr_creativity", package = "mtscr")
mtscr_score(mtscr_creativity, id, item, SemDis_MEAN, top = 1:2)
#> # A tibble: 149 × 3
#> id .creativity_score_top1 .creativity_score_top2
#> <chr> <dbl> <dbl>
#> 1 84176 0.142 0.0681
#> 2 84177 -0.508 -0.494
#> 3 84178 -0.0733 -0.0995
#> 4 84188 0.529 0.527
#> 5 84193 -0.299 -0.350
#> 6 84206 -0.312 -0.301
#> 7 84211 -0.0464 0.0356
#> 8 84226 0.238 0.210
#> 9 84228 0.137 0.139
#> 10 84236 0.459 0.422
#> # ℹ 139 more rows
mtscr_score() does everything automatically. You can also use mtscr_prepare() to get your data prepared for modelling by hand and mtscr_model() to get the model object. See the functions’ documentation for more details.
The model can be summarised to obtain the parameters and reliability estimates.
mtscr_model(mtscr_creativity, id, item, SemDis_MEAN, top = 1:3) |>
mtscr_model_summary()
#> # A tibble: 3 × 10
#> model nobs sigma logLik AIC BIC deviance df.residual emp_rel FDI
#> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 top1 4585 0.736 -5298. 10657. 10850. 2383. 4555 0.877 0.936
#> 2 top2 4585 0.767 -5472. 11003. 11196. 2597. 4555 0.892 0.944
#> 3 top3 4585 0.825 -5777. 11613. 11806. 3024. 4555 0.896 0.947
Graphical User Interface
This package includes a Shiny app which can be used as a GUI. You can find “mtscr GUI” option in RStudio’s Addins menu. Alternatively execute mtscr_app() to run it.
Try web based version here!
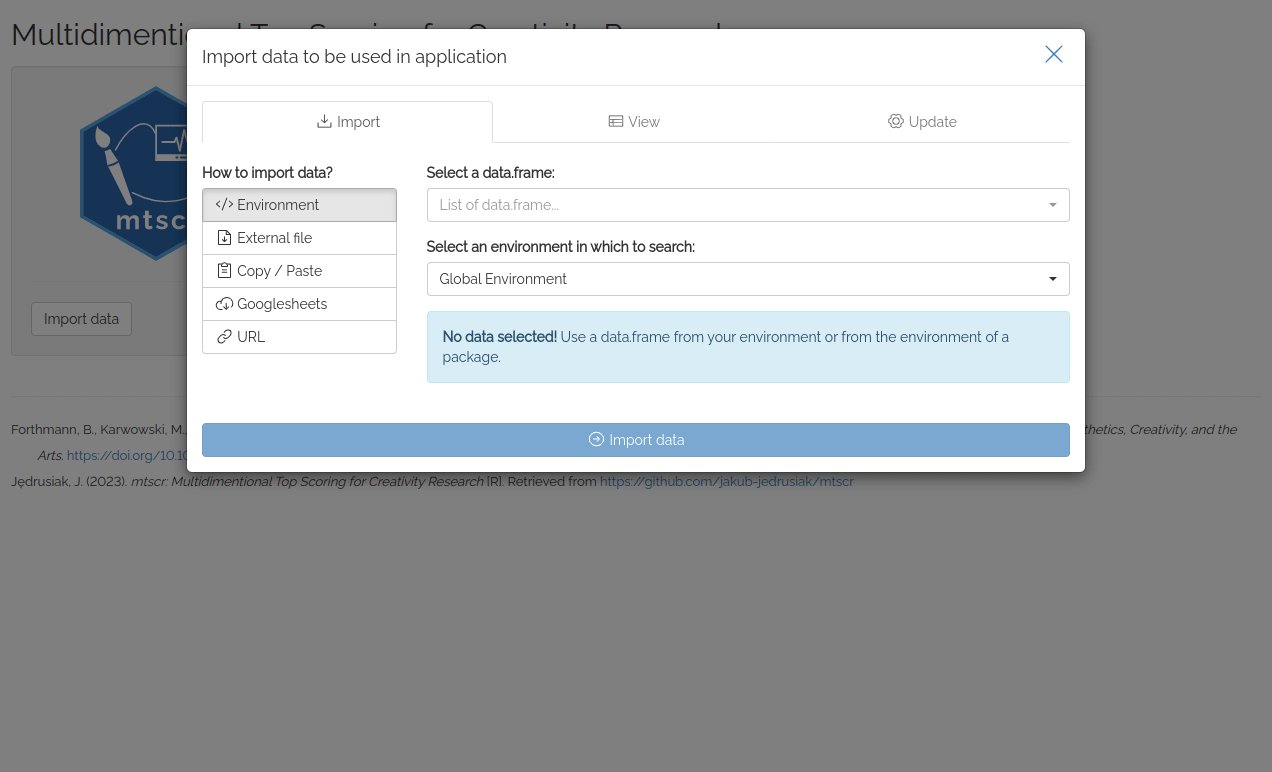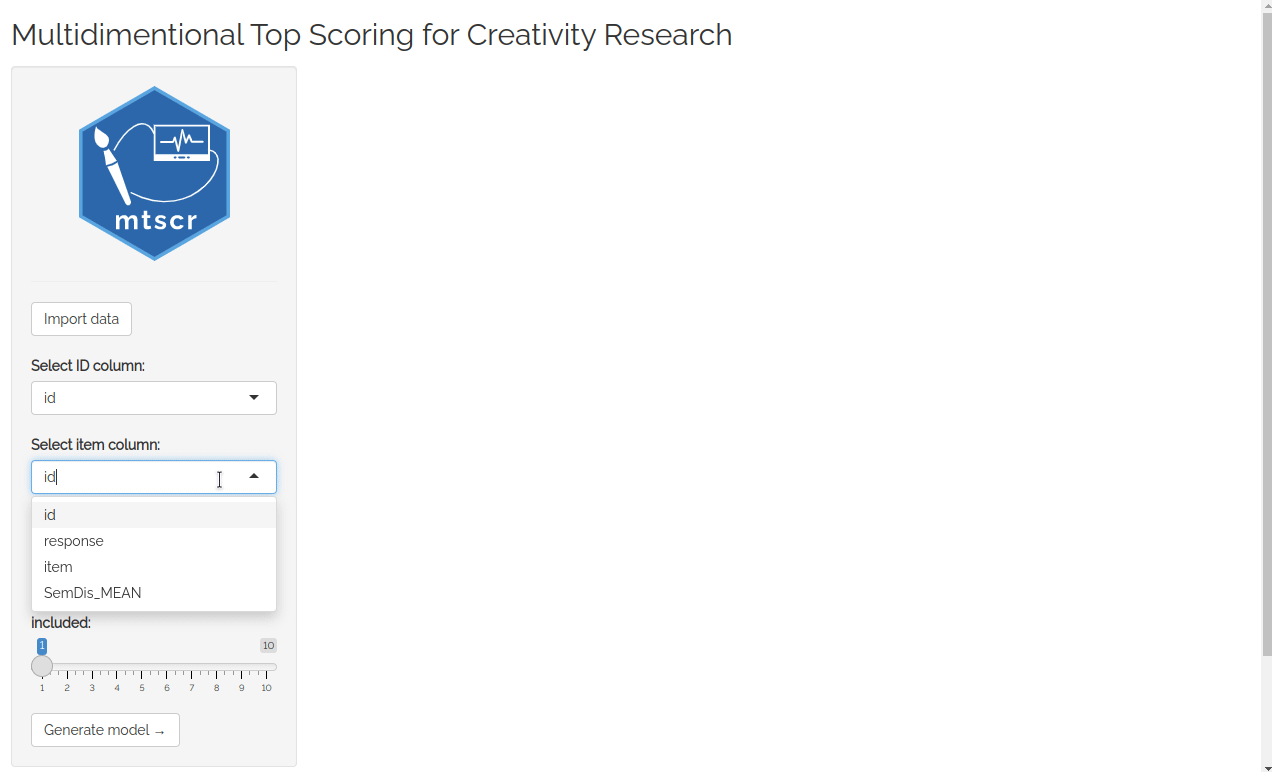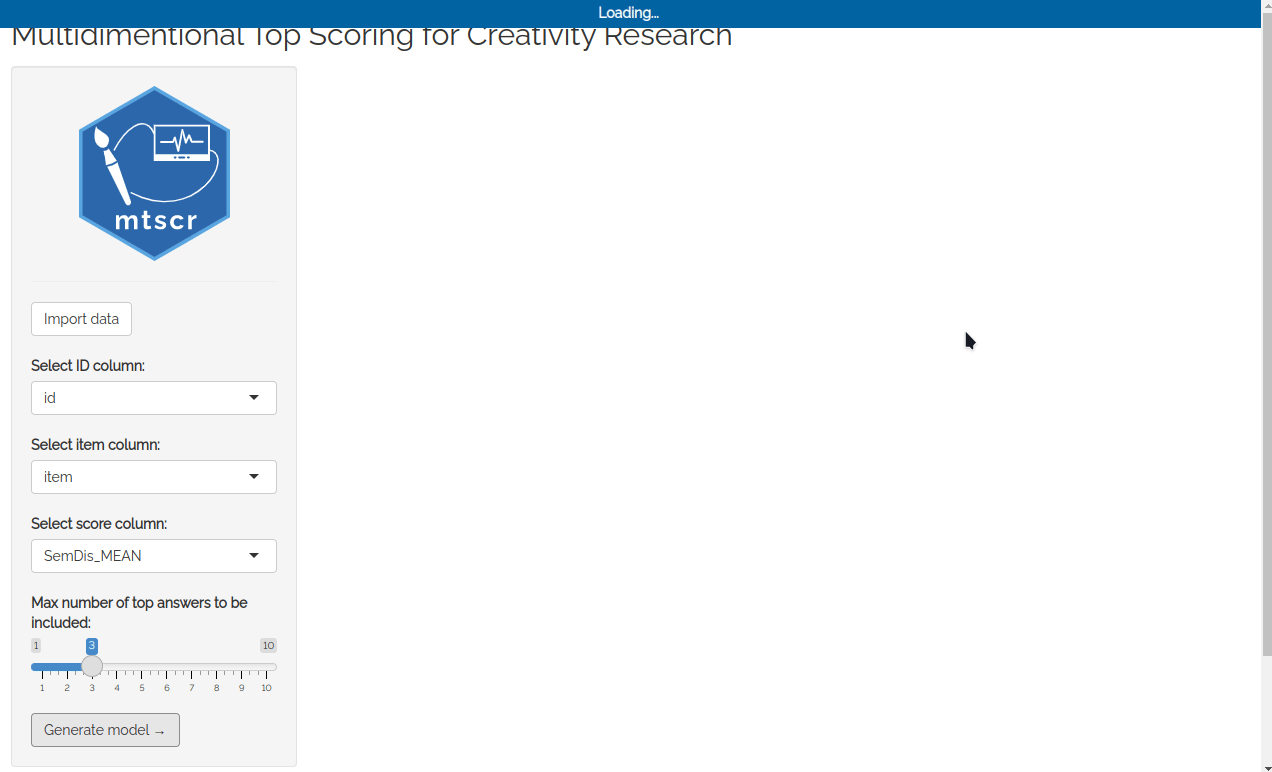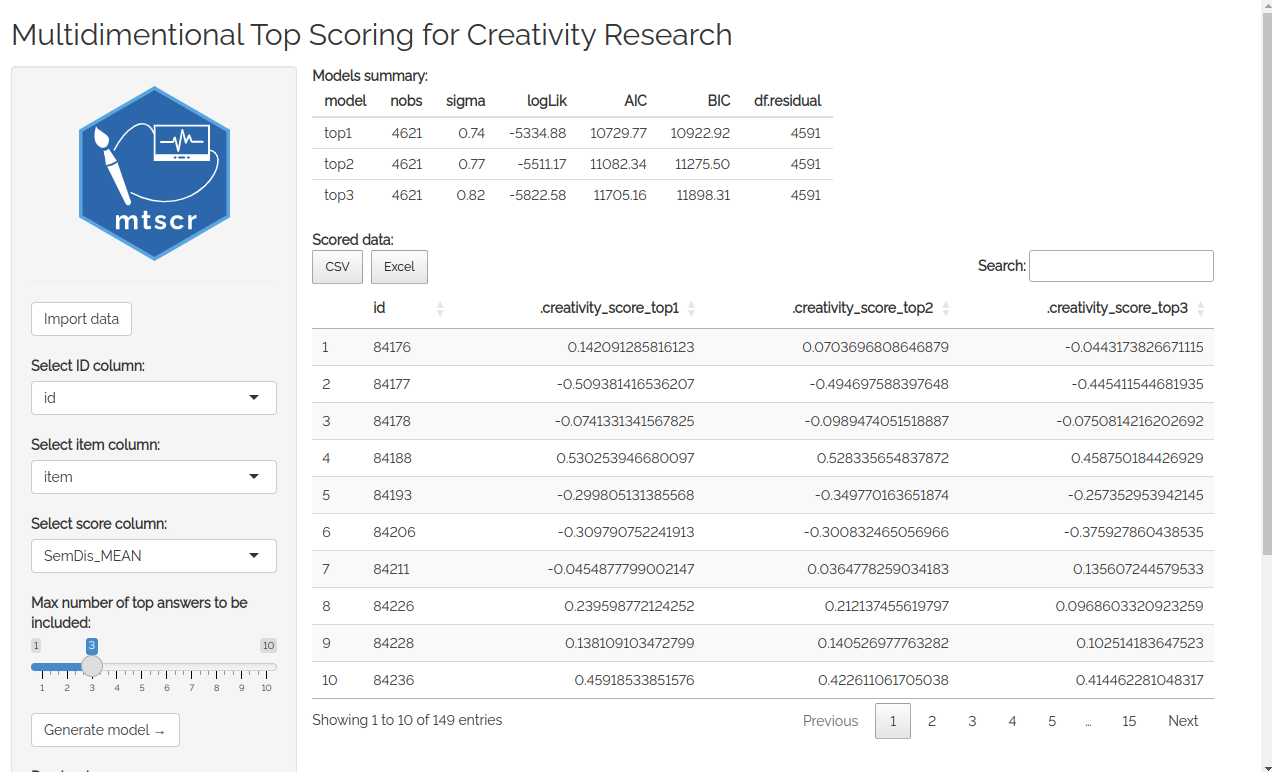
First thing you see after running the app is datamods window for importing your data. You can use the data already loaded in your environment or any other option. Then you’ll see four dropdown lists used to choose arguments for mtscr_model() and mtscr_score() functions. Consult these functions’ documentation for more details (execute ?mtscr_score in the console). When the parameters are chosen, click “Generate model” button. After a while (up to a dozen or so seconds) models’ parameters and are shown along with a scored dataframe.
You can download your data as a .csv or an .xlsx file using buttons in the sidebar. You can either download the scores only (i.e. the dataframe you see displayed) or your whole data with scores columns added.
For testing purposes, you may use mtscr_creativity dataframe. In the importing window change “Global Environment” to “mtscr” and our dataframe should appear in the upper dropdown list. Use id for the ID column, item for the item column and SemDis_MEAN for the score column.
Contact
Correspondence concerning the meritorical side of these solutions should be addressed to Boris Forthmann, Institute of Psychology, University of Münster, Fliednerstrasse 21, 48149 Münster, Germany. Email: [email protected].
The maintainer of the R package is Jakub Jędrusiak and the technical concerns should be directed to him. Well, me. Best way is to open a discussion on GitHub. Technical difficulties may deserve an issue.

