Description
Beautiful Graph Drawing.
Description
A graph visualization engine that emphasizes on aesthetics at the same time providing default parameters that yield out-of-the-box-nice visualizations. The package is built on top of 'The Grid Graphics Package' and seamlessly work with 'igraph' and 'network' objects.
README.md
netplot
An alternative graph visualization tool that emphasizes aesthetics, providing default parameters that deliver out-of-the-box lovely visualizations.
Some features:
- Auto-scaling of vertices using sizes relative to the plotting device.
- Embedded edge color mixer.
- True curved edges drawing.
- User-defined edge curvature.
- Nicer vertex frame color.
- Better use of space-filling the plotting device.
The package uses the grid plotting system (just like ggplot2).
Installation
You can install the released version of netplot from CRAN with:
install.packages("netplot")
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("USCCANA/netplot")
Example
This is a basic example which shows you how to solve a common problem:
library(igraph)
#>
#> Attaching package: 'igraph'
#> The following objects are masked from 'package:stats':
#>
#> decompose, spectrum
#> The following object is masked from 'package:base':
#>
#> union
library(netplot)
#> Loading required package: grid
#>
#> Attaching package: 'netplot'
#> The following object is masked from 'package:igraph':
#>
#> ego
set.seed(1)
data("UKfaculty", package = "igraphdata")
l <- layout_with_fr(UKfaculty)
#> This graph was created by an old(er) igraph version.
#> Call upgrade_graph() on it to use with the current igraph version
#> For now we convert it on the fly...
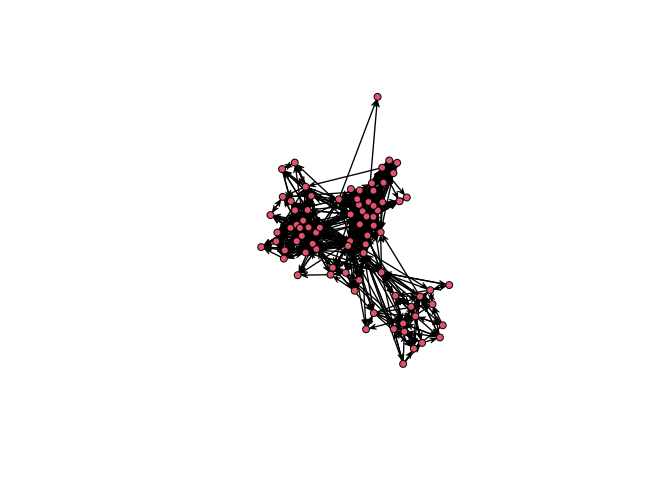
plot(UKfaculty, layout = l) # ala igraph
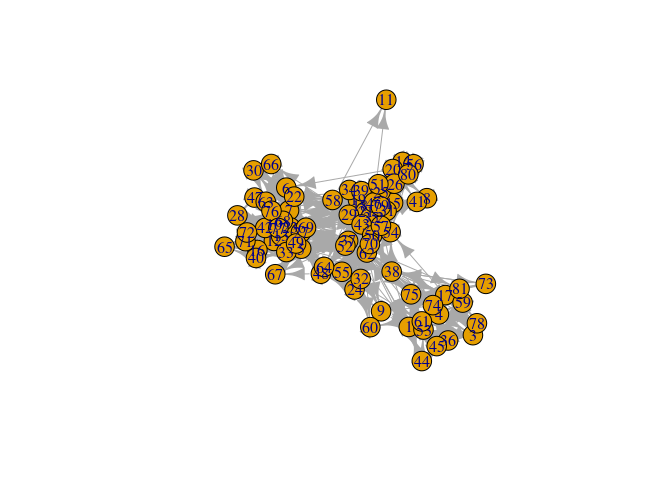
V(UKfaculty)$ss <- runif(vcount(UKfaculty))
nplot(UKfaculty, layout = l) # ala netplot
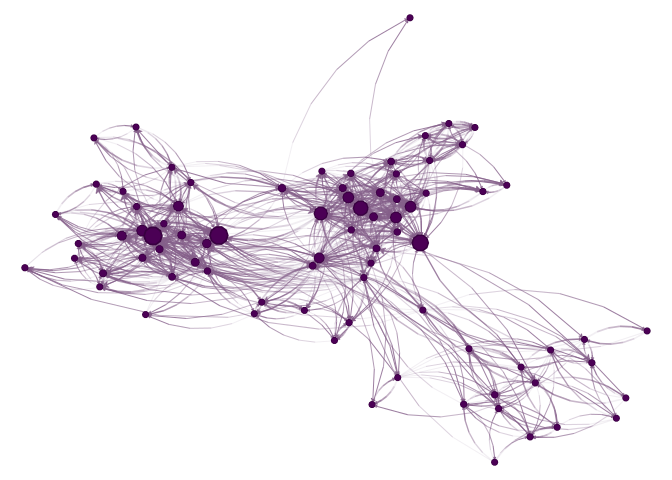
sna::gplot(intergraph::asNetwork(UKfaculty), coord=l)
UKfaculty
# Random names
set.seed(1)
nam <- sample(babynames::babynames$name, vcount(UKfaculty))
ans <- nplot(
UKfaculty,
layout = l,
vertex.color = ~ Group,
vertex.nsides = ~ Group,
vertex.label = nam,
vertex.size.range = c(.01, .03, 4),
bg.col = "transparent",
vertex.label.show = .25,
vertex.label.range = c(10, 25),
edge.width.range = c(1, 4, 5),
vertex.label.fontfamily = "sans"
)
# Plot it!
ans

Starting version 0.2-0, we can use gradients!
ans |>
set_vertex_gpar(
element = "core",
fill = lapply(get_vertex_gpar(ans, "frame", "col")$col, \(i) {
radialGradient(c("white", i), cx1=.8, cy1=.8, r1=0)
}))

USairports
# Loading the data
data(USairports, package="igraphdata")
# Generating a layout naively
layout <- V(USairports)$Position
#> This graph was created by an old(er) igraph version.
#> Call upgrade_graph() on it to use with the current igraph version
#> For now we convert it on the fly...
layout <- do.call(rbind, lapply(layout, function(x) strsplit(x, " ")[[1]]))
layout[] <- stringr::str_remove(layout, "^[a-zA-Z]+")
layout <- matrix(as.numeric(layout[]), ncol=2)
# Some missingness
layout[which(!complete.cases(layout)), ] <- apply(layout, 2, mean, na.rm=TRUE)
# Have to rotate it (it doesn't matter the origin)
layout <- netplot:::rotate(layout, c(0,0), pi/2)
# Simplifying the network
net <- simplify(USairports, edge.attr.comb = list(
weight = "sum",
name = "concat",
Passengers = "sum",
"ignore"
))
# Pretty graph
nplot(
net,
layout = layout,
edge.width = ~ Passengers,
edge.color = ~
ego(col = "white", alpha = 0) +
alter(col = "yellow", alpha = .75),
skip.vertex = TRUE,
skip.arrows = TRUE,
edge.width.range = c(.75, 4, 4),
bg.col = "black",
edge.line.breaks = 10
)




