Noise Quantification in High Throughput Sequencing Output.
noisyR: Enhancing biological signal in sequencing datasets by characterising random technical noise
High-throughput sequencing enables an unprecedented resolution in transcript quantification, at the cost of magnifying the impact of technical noise. The consistent reduction of random background noise to capture functionally meaningful biological signals is still challenging. Intrinsic sequencing variability introducing low-level expression variations can obscure patterns in downstream analyses.
The noisyR package comprises an end-to-end pipeline for quantifying and removing technical noise from HTS datasets. The three main pipeline steps are [i] similarity calculation across samples, [ii] noise quantification, and [iii] noise removal; each step can be finely tuned using hyperparameters; optimal, data-driven values for these parameters are also determined.
Preprint: https://www.biorxiv.org/content/10.1101/2021.01.17.427026v2
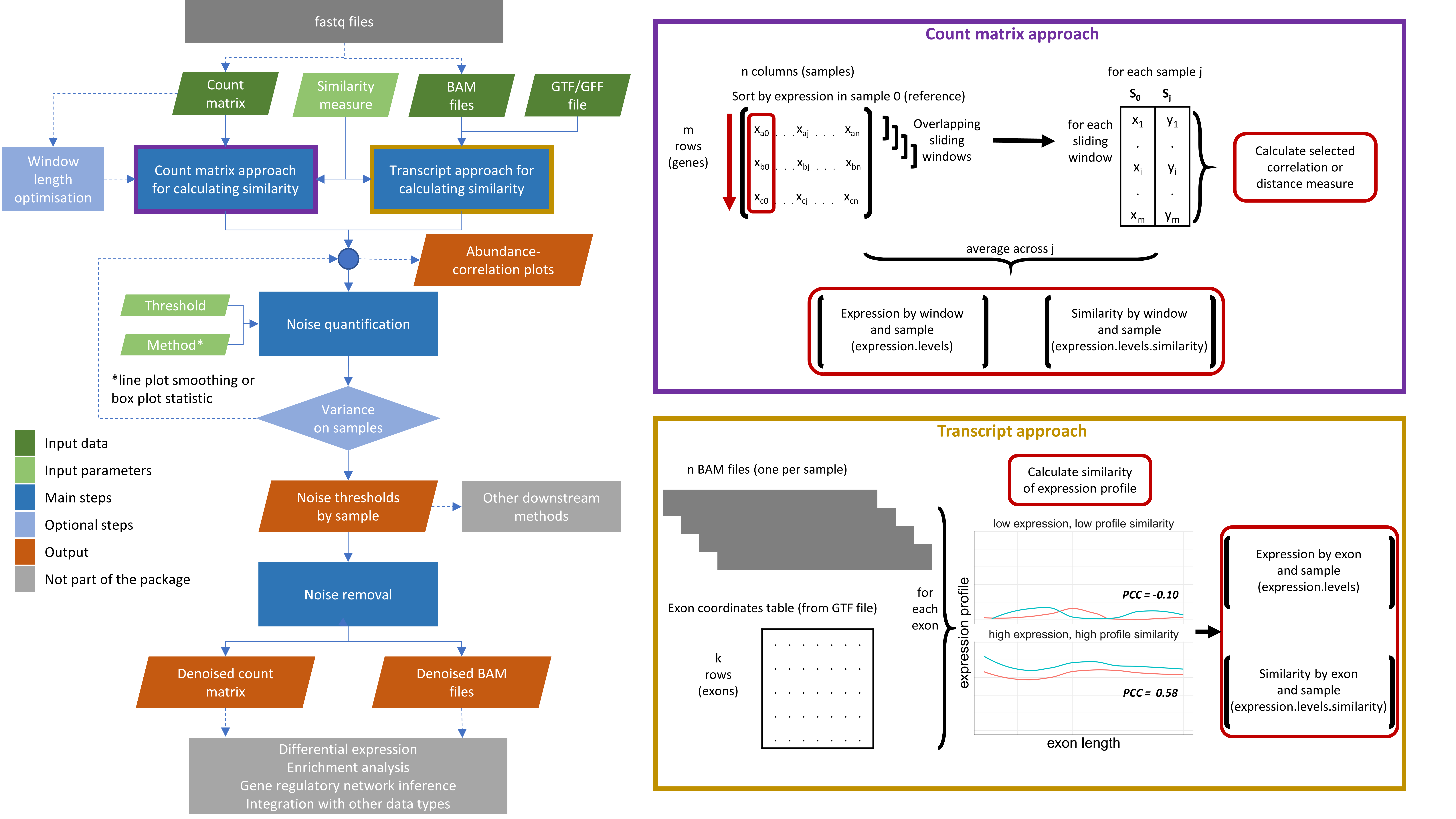
Workflow diagram of the noisyr pipeline
Similarity calculation across samples
For the sample-similarity calculation, two approaches are available:
- The count matrix approach uses the original, un-normalised expression matrix, as provided after alignment and feature quantification; each sample is processed individually, only the relative expressions across samples are compared. Relying on the hypothesis that the majority of genes are not DE, most of the evaluations are expected to point towards a high similarity across samples. Choosing from a collection of >45 similarity metrics, users can select a measure to assess the localised consistency in expression across samples. A sliding window approach is used to compare the similarity of ranks or abundances for the selected features between samples. The window length is a hyperparameter, which can be user-defined or inferred from the data.
- The transcript approach uses as input the alignment files derived from read-mappers (in BAM format). For each sample and each exon, the point-to-point similarity of expression across the transcript is calculated across samples in a pairwise all-versus-all comparison.
The output formats for the two approaches correspond to their inputs: a denoised count matrix for the count matrix approach, and denoised BAM files for the transcript approach.
Main functions: calculate_expression_similarity_counts(), calculate_expression_similarity_transcript()
Supporting functions: get_methods_correlation_distance(), optimise_window_length(), calculate_expression_profile()
Input preparation functions: cast_matrix_to_numeric(), cast_gtf_to_genes()
Noise quantification
The noise quantification step uses the expression-similarity relation calculated in step i to determine the noise threshold, representing the level below which the gene expression is considered noisy.
For example, if a similarity threshold is used as input then the corresponding expression from a (smoothed) expression-similarity line plot is selected as the noise threshold for each sample. The shape of the distribution can vary across experiments; we provide functionality for different thresholds and recommend the choice of the one that results in the lowest variance in the noise thresholds across samples.
Options for smoothing, or summarising the observations in a box plot and selecting the minimum abundance for which the interquartile range (or median, or 5-95% range) is consistently above the similarity threshold are also available. As a general rule (due to the number of observations), we recommend using the smoothing with the count matrix approach, and the boxplot representation with the transcript option.
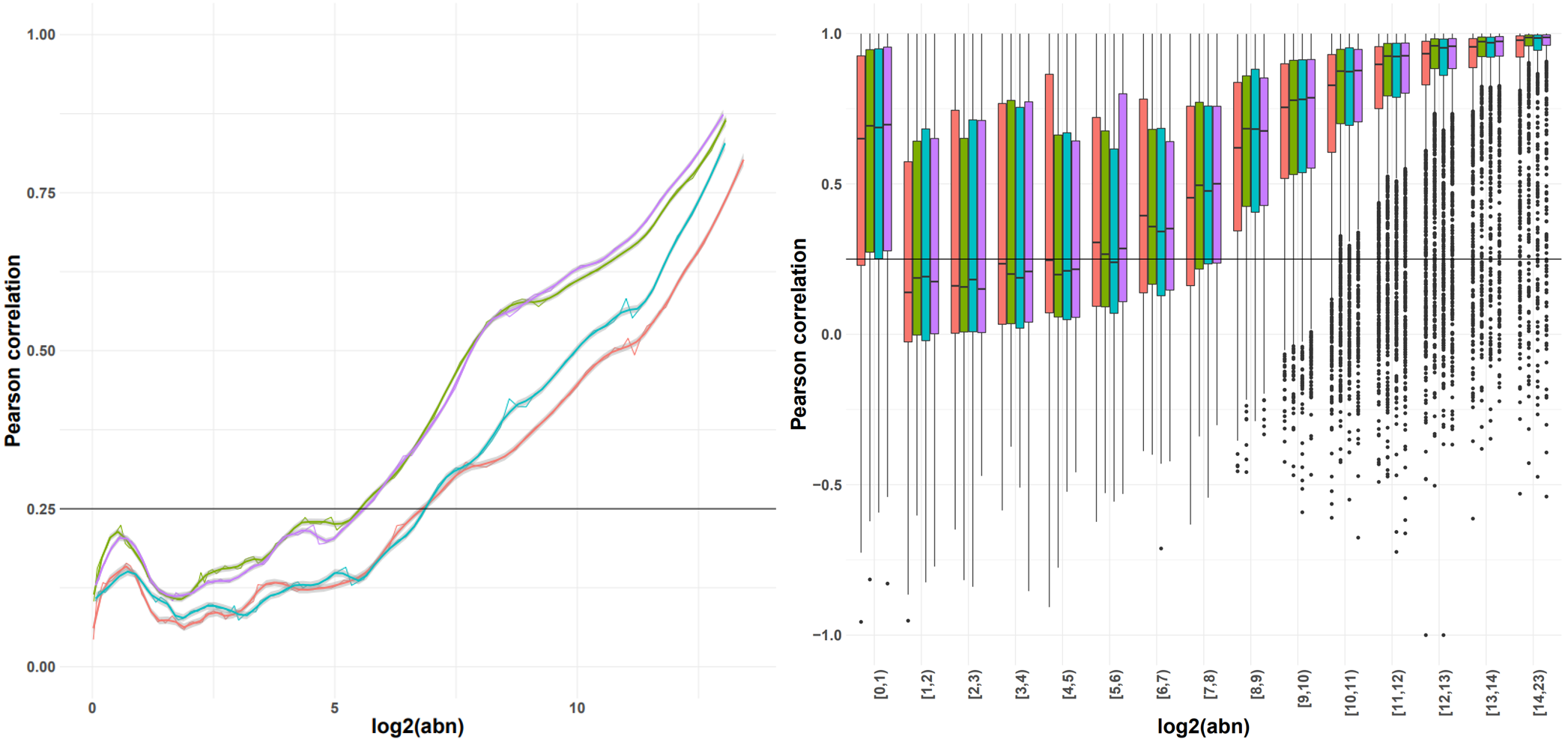
Indicative plots of the Pearson correlation calculated on windows of increasing average abundance for the count matrix-based noise removal approach (left) and per exon for the transcript-based noise removal approach (right).
Main function: calculate_noise_threshold_base()
Supporting functions: get_methods_calculate_noise_threshold(), calculate_first_minimum_density()
Visualisation functions: plot_expression_similarity(), calculate_noise_threshold_method_statistics()
Noise removal
The third step uses the noise threshold calculated in step ii to remove noise from the count matrix or BAM files.
- For the count matrix approach, genes whose expression is below the noise thresholds for every sample are removed and the average noise threshold is calculated and added to every entry. This ensures that the fold-changes observed by downstream analyses are not biased by low expression, while still preserving the structure and relative expression levels in the data.
- For the transcript approach, genes are removed from the BAM files if the expression of all their exons is below the noise thresholds for every sample. The removal is done at gene level to avoid scenarios that are not biologically possible.
Main functions: remove_noise_from_matrix(), remove_noise_from_bams()
Required packages
CRAN (use install.packages())
- utils
- grDevices
- tibble
- dplyr
- magrittr
- ggplot2
- philentropy
- doParallel
- foreach
Bioconductor (use BiocManager::install())
- preprocessCore
- IRanges
- GenomicRanges
- Rsamtools.